| Prev | ICM User's Guide 10.13 Docking Templates | Next |
You can use either substructure or Atomic Property Field Templates for docking.
- Setup the ligand and receptor.
- File/Open a 3D template structure and place in the pocket.
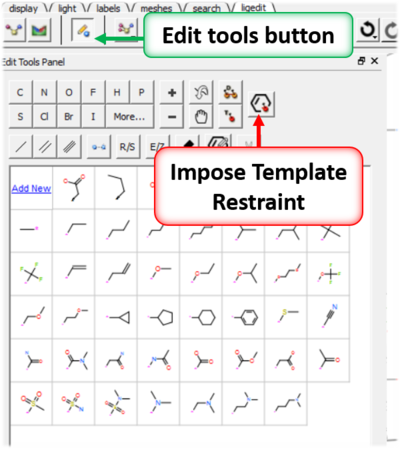
- Click on the Edit Tools panel button.
- Click on the Impose Template Restraint button.
- Select the template object from the drop down menu and choose whether you wish to impose substructure or 3D pharmacophore APF template.
- A table will be displayed called DOCK_RESTRAINTS where you can turn the template on or off.
| Prev Distance Restraint | Home Up | Next Flexible Groups |