| Prev | ICM User's Guide 9.1 Homology Modeling | Next |
[ Homology Modeling Introduction | Quick Model | Full Model Builder | Model Editor ]
| Available in the following product(s): Homology |
9.1.1 Homology Modeling Introduction |
[ Modeling Options | Getting Started | Older Versions ]
9.1.1.1 Modeling Options |
There are three main modeling options to choose from:
- Quick Model. This option will provide a model by placement of the aligned polypeptide chain onto the template structure. The method is fast and is ideal if you want a quick model for visualization purposes.
- Full Model Builder. This option will give you a fully refined model and can take many hours to complete the model building process. There are options to fully refine side-chains, refine side-chains and loops or undertake a full refinement. The refinement is undertaken using the ICM BPMC modeling method in the ICM force field which includes surface terms, electrostatics with the boundary element solution of the Poisson equation and side chain entropy terms.
- Multi-Template Model Editor. This option is ideal if you want more control over your model - particularly in loop regions or if you want to use multiple templates. It provides an interface to add new templates to a model and browse through multiple conformations of loop regions. It also provides an interactive visualization interface to see how well the residues in your model falls into the Ramachandran Plot and how good the omega angles are.
9.1.1.2 Homology Modeling Getting Started |
The items you need to begin making an Homology Model are:
- A template structure from the PDB converted into an ICM object.
- Extract the sequence of the template structure from the PDB file.
- The sequence of your query molecule (e.g. from UniProt or FASTA).
- An alignment (see alignment section) of your query sequence against the template sequence from the PDB.
Your graphical user interface window should look something like this:
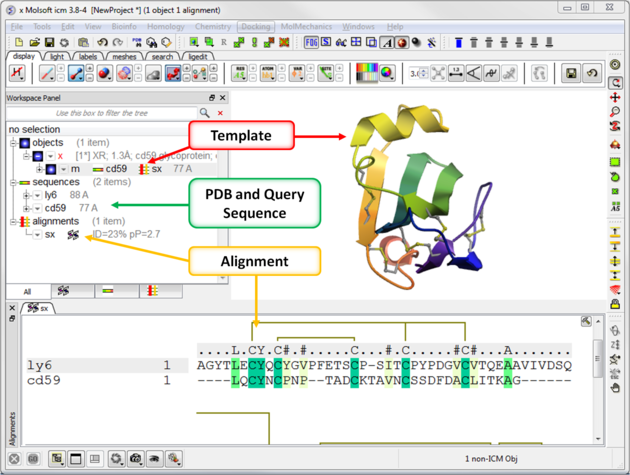
9.1.1.3 Older Versions of Homology Modeling in ICM |
If you are using a version of ICM 3.8-3 or earlier you will see different options in the Homology Menu. We recommend updating your ICM version to 3.8-4 or newer at www.molsoft.com/support If you cannot update your ICM version then Homology/Build model is similar to Quick Model, Homology
9.1.2 Quick Model |
[ Single Chain | Multi Chain ]
9.1.2.1 Single Chain from Sequence |
To build a 'Quick' homology model from a single chain:
- Check that you have your query sequence, template structure and alignment loaded in ICM.
- Click on the 'Homology' menu at the top of the graphical user interface.
- Select Quick Model and a dialog box will be displayed as shown below.
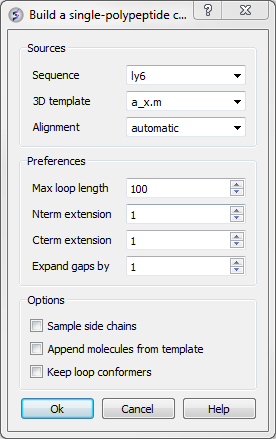
This data entry box is split into 3 sections, the first is 'sources' where you need to specify your query sequence, template and alignment. The second section is called 'preferences where penalty information for the model needs to be entered and the third section is called 'Options'. Each will be described in detail below.
To construct your model follow these steps:
- Enter the name of your QUERY (ie the sequence of the model you wish to build - NOT the template) sequence in the data entry box labeled Sequence.If you click on the arrow next to this box a list of sequences loaded in ICM will be displayed. The names of the sequences are also listed in the workspace panel on the left of the graphical user interface.
- Enter the name of your template structure in the '3D template' data entry box. Once again the name of your template structure can be found by clicking the down arrow or in the workspace panel.
- Enter the name of your alignment in the 'Alignment' data entry box.
You could build your model now as ICM has enough information but it may be wise to take a look at some of the preferences and change them accordingly. However in most cases the default values provided are sufficient to produce a good quality model.
To change the preferences either type the number you wish or use the up and down arrows next to the data entry boxes.
- Max loop length (default= 999) - loops longer than this value are not modeled
- Nterm extension (default=1) - the maximal length of the N-terminal model sequence which extends beyond the template
- Cterm extension (default=1) - the maximal length of the C-terminal model sequence which extends beyond the template
- Expand gaps by (default=1) - additional widening of the gaps in the alignment. End gaps are not expanded
Now all you need to do to build your model is to select some options. The options are:
- Sample side chains - performs monte-carlo optimization on the side chains
- Append molecules from template - this option will transfer all hetatoms and molecules in the template structure to your model.
- Keep loop conformers - this option will save alternative loop conformations in the ICM workspace.
To build your model:
- Click OK
Once your model is built a new object containing your model will be available in the ICM workspace panel (left hand side).
9.1.2.2 Multi-Chain from Linked Alignment |
To make a multi-chain model you must first link all the chains to a template via an alignment. For example, in an antibody you have light chains and heavy chains so you would make two alignments with the chains in the template structure.
To do this:
- Read in the template PDB structure.
- Read in the query sequence.
- Extract the sequence from the chains in the PDB you wish to model.
- Make alignments of the query and chain sequences.
- Go to Homology/Quick Model/Multi-Chain from Linked Alignment
- Enter the name of the template structure.
- Enter a name for the model you are going to build .
- Click "sample side-chains" if required.
- When the model is ready it will be displayed in the ICM Workspace.
9.1.3 Full Model Builder |
[ Multi Chain ]
The Full Model Builder will give you a fully refined model and can take many hours to complete the model building process. There are options to fully refine side-chains, refine side-chains and loops or undertake a full refinement. The refinement is undertaken using the ICM BPMC modeling method in the ICM force field which includes surface terms, electrostatics with the boundary element solution of the Poisson equation and side chain entropy terms.
- Check that you have your query sequence, template structure and alignment loaded in ICM.
- Click on the 'Homology' menu at the top of the graphical user interface.
- Select Full Model Builder and a dialog box will be displayed as shown below.
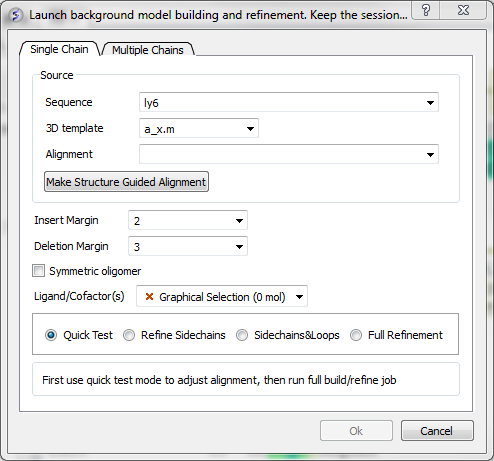
- Enter the name of your QUERY (ie the sequence of the model you wish to build - NOT the template) sequence in the data entry box labeled Sequence.If you click on the arrow next to this box a list of sequences loaded in ICM will be displayed. The names of the sequences are also listed in the workspace panel on the left of the graphical user interface.
- Enter the name of your template structure in the '3D template' data entry box. Once again the name of your template structure can be found by clicking the down arrow or in the workspace panel.
- Enter the name of your alignment in the 'Alignment' data entry box.
- Enter a value for Insert Margin. Insert margin is the number of residues that will be untethered around an insertion in the modeled sequence in sequence-structure alignment.
- Enter a value for Deletion Margin. Delete margin is the number of residues around the deletion in the query (insertion in the template) that are untethered.
- Select oligomeric if you want to build a model of an oligomer provided in the template structure.
- You can choose to include Ligands and Co-factors by selecting them using the orange selections
- First use Quick Test to check the model and make any necessary adjustments to the alignment. When you are happy, proceed to refine-side chains, or refine side-chains and loops, or full refinement.
9.1.3.1 Full Model of Multi-Chain from Linked Alignment |
To make a multi-chain model you must first link all the chains to a template via an alignment. For example, in an antibody you have light chains and heavy chains so you would make two alignments with the chains in the template structure.
To do this:
- Read in the template PDB structure.
- Convert the PDB file into an ICM object.
- Read in the query sequence.
- Extract the sequence from the chains in the PDB you wish to model.
- Make alignments of the query and chain sequences.
- Go to Homology/Full Model Builder
- Click on the Multiple Chain tab.
- Use the check boxes in the sequence drop down button to select multiple sequences for the chains you are modeling.
- Use the check boxes in the alignment drop down button to select the sequence alignment for each chain.
9.1.4 Multi-Template Model Editor |
[ Modeler's View | Interactive Loop | Multi Template ]
| Available in the following product(s): ICM-Homology |
What you need before you can use the Multi-Template Model Editor:
Build a New Model
- A template structure from the PDB converted (See convert object) into an ICM object.
- A sequence file of your query sequence.
You can use an alignment you have constructed yourself or allow ICM to generate one (referred to as automatic)
- An alignment (see alignment section) of your query sequence against the template structure from the PDB. This is a *.ali file in ICM.
- Homology/Multi-Template Model Editor
- Enter the name of your loaded sequence.
- Enter the name of your loaded 3D template
- Enter the name of your alignment or allow ICM to generate an alignment by selecting automatic
- Check whether you wish ICM to sample the loop regions of your model or not
A Interactive Modeler's View of the alignment will then be displayed.
Pre-Existing Model
If you have built a model using Quick Model or the Full Model Builder you can edit this model in the editor by:
- Homology/Multi-Template Model Editor/Setup from Existing Model
- Enter the name of the Model and The Template you wish to use for multi-template modeling.
A Interactive Modeler's View of the alignment will then be displayed.
9.1.4.1 Modeler's View |
Once you have made your interactive model your graphical user interface should look something like this:
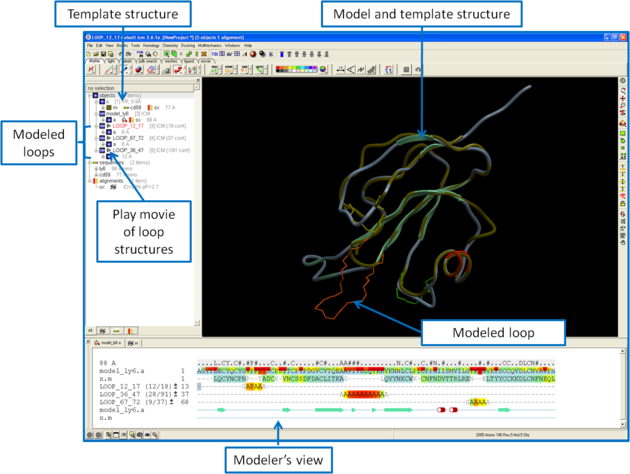
What do all the elements of the Modeler's View mean?
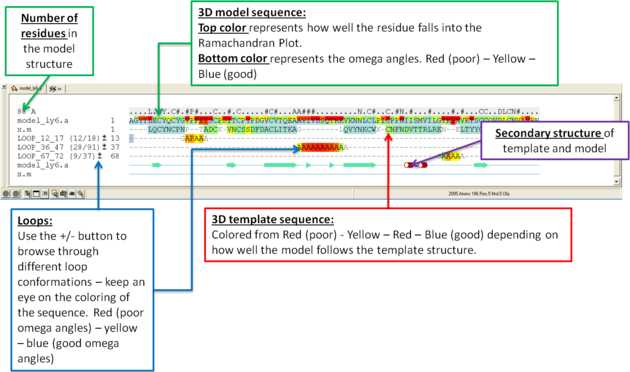
9.1.4.2 Interactive Loop Modeling |
You can browse a number of different loop conformations by clicking on the +/- button next to the loop in the modeler view's window. Keep an eye on the colors of the loop residues in the alignment - they are colored red (poor) -yellow - blue (good) depending on how good the omega angles are. The residues in the loops are represented by Alanines for every residue other than Glycine or Proline.
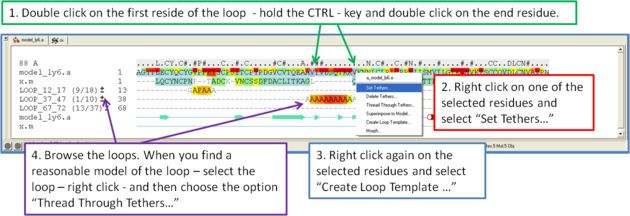
If you want to remodel the loop.
- Set tethers at the residue at the start and end of the loop. To set a tether double click on the two residues whilst holding down the CTRL key. Right click and select Set tethers. Selecthow you want to tether to the template either by alignment, residue numbering or selection. Select the alignment - automatic represents the Modeler's View. Select the Template Molecule which is the object you built the model on.
- Select the loop you wish to model including the tethered residues at the start and end of the loop and then right click and select Create Loop Template
- Once the simulation has finished you can once again browse the solutions by clicking on the +/- button next to the loop in the modeler view's window.
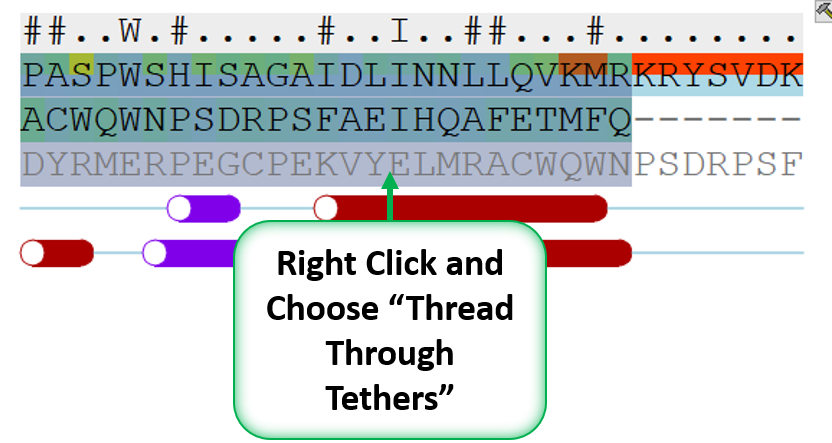
- When you have identified a reasonable conformation of the loop you can re-thread it onto the model structure by selecting the loop region in the Modeler's View. You can do this by clicking and dragging over the loop. Right click and select Thread Through Tethers.
9.1.4.3 Multi Template Model |
You can use twp or more templates to build a model. In the example below we have two template structures A.pdb and B.pdb and our query sequence.
- Read in PDB structures A.pdb and B.pdb
- Read in the query sequence for your model.
- Extract the sequence from each template structure A.pdb and B.pdb (right click on the chain in the icm workspace and choose extract sequence)
- Make two sequence alignments A.pdb vs query and another separate alignment B.pdb vs query. If you have more than two templates you will need to make a separate alignment for each one.
- First make an initial model of query sequence based on A.pdb and setup the modeler's view alignment (Homology/Multi-Template Model Editor/Setup from Sequence and Template). Enter the data as shown below and click OK.
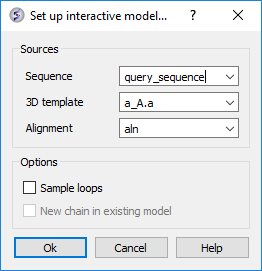
- You will then see the Modeler's View Alignment and a Model in the ICM Workspace.
- Select in the Modeler's View Alignment alignment (left click and drag) where you would like to model the structure of template B.pdb onto your current model.
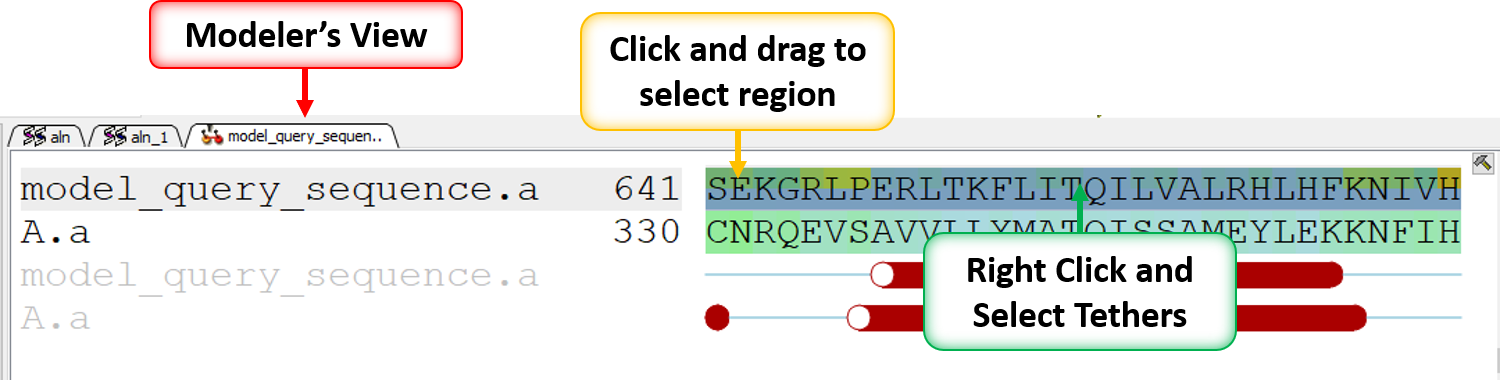
- Right click on the alignment selection and choose "Set Tethers". Choose "Use alignment" option and then select the alignment of query vs B.pdb and the template structure B.pdb in the drop down window Click OK.
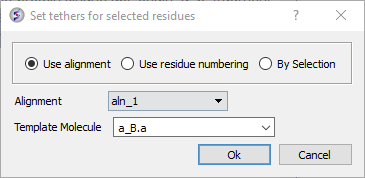
- Once the tethers have been made go back to the selection you made in the Modeler's view alignment and then right click again and choose "Thread Through Tethers" and the model will be built.
| Prev Protein Modeling | Home Up | Next Loop Modeling |