ICM sequences and objects may contain specific information about local sequence features, such as location of binding sites, active sites, effects of mutations, etc. This information is stored in the feature tables in protein sequence and structure entries. We have combined all the annotations, transferred them structures and stored them in a compact form for immediate access and fast visualization. Many categories of functional residue sites are accessble and can be separately analyzed and visualized.
We use one letter code (the second column)†to specify the site type.
Some details about XPDB:
- Compact, requires one DVD-ROM (2.9Gb).
- Includes a more compact version (0.5Gb) XPDB-Mini for instant local browsing.
- Correct bond types for ligands (HET-residues) to enable conversion and docking.
- XPDB†contains all PDB entries at the time of release in the parsed binary format.
- XPDB entries contain information about SNPs assigned to residues of 3D model.
- Contains functional annotation transferred from sequence databases by alignment and empirical rules.
- Can be viewed by ICM-Browser (free), ICM-Browser-Pro and ICM-Pro.
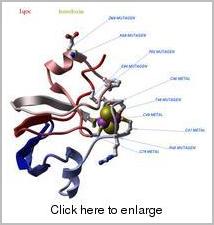
An example XPDB image generated by ICM-Browser.