Google Search: Keyword Search:
| Prev | ICM User's Guide 14 ICM in KNIME | Next |
| Video - Installation | Video - MolCalcProp Node |
KNIME (Konstanz Information Miner) is a user-friendly and comprehensive open-source data integration, processing, analysis, and exploration platform. Currently we have KNIME nodes for the following ICM functions:
Available Nodes
Docking and Screening
- MolIcmDockPrep - Prepare receptor and ligand for docking. Covalent docking support for MolIcmDockPrep node (ICM 3.8-1 or higher is required)
- MolIcmDock - Ligand Docking and Screening (_dockScan and _dockProjPrep). Recently we have added "Output Type" for MolIcmDock node. It allows to choose between SDF and interactive ICM HitList which can be viewed using the MolView node.
- MolScreen - MolScreen is a set of high quality 2D fingerprint and 3D pharmacophore models for a broad range of pharmacology and toxicology targets.
Cheminformatics
- MolConverTo3D - Chemical 2d to 3d conversion.
- MolConfGen - Conformation generator (_confGen).
- MolAPFSuperimpose - Flexible APF superposition (_chemSuper).
- MolCalcProp - Allows to calculate various chemical properties and desciptors (e.g. Calculate MolArea, MolWeight, InChi...). For a complete list of available functions please click here.
- MolStandardize - Chemical Table/Spreadsheet standardization - remove salt, remove hydrogen, standardize groups, apply 2D depiction, assign formal charges.
- MolUnique - removes duplicate molecules.
- MolCenters - Performs chemical clustering and selects a specified number of representatives.
- MolCluster - Cluster either by fingerprints or by numerical column selection.
- MolView - Connect this node to the output of other Molsoft nodes. (e.g. MolScreen, MolIcmDock...).
To use ICM in KNIME
- Download KNIME (Analytics Platform) from www.knime.org
- In KNIME go to File/Install KNIME extensions and search for and select the KNIME Base Chemisty Types and Nodes. When you do this you will see a Chemistry directory in the Node Repository.
- Download MolSoft's KNIME Java (see instructions below). Note, that if you have an old version of Molsoft Nodes installed manually you need to remove the jar file and restart KNIME with "-clean" option.
MolSoft Knime
- To enable automatic update of molsoft nodes: go to File/Preferences. Open Install/Update - Available Software Sites, Click on "Add..." button and enter information below:
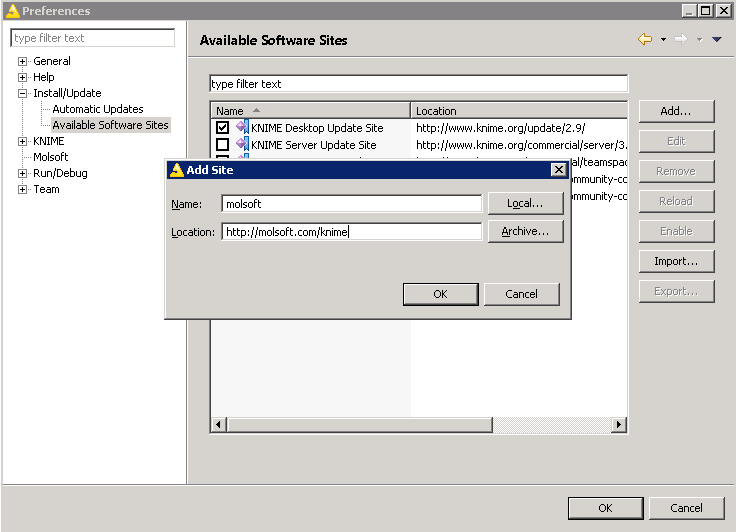
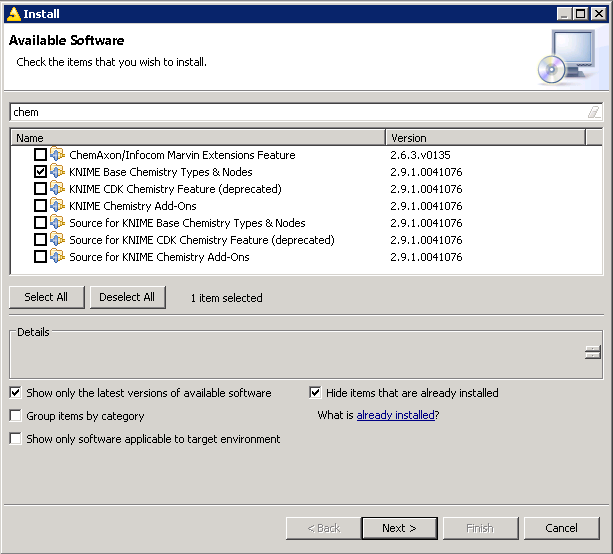
- Then, goto "File/Install update extensions", uncheck "Group items by category" If you don't have "Base Chemistry" package installed, then install it first (type 'chem' to locate package)
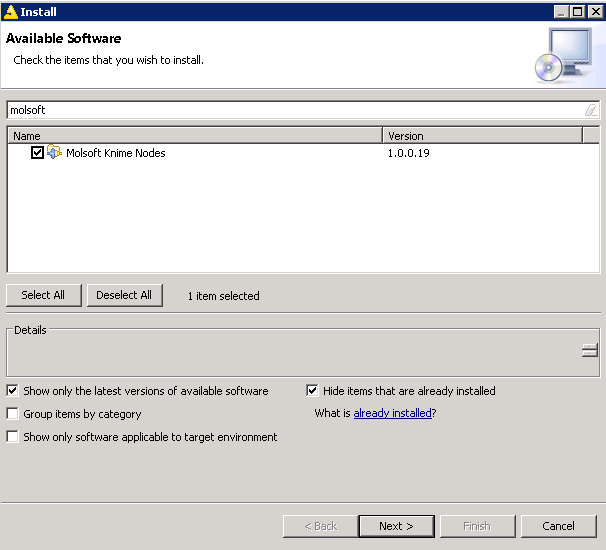
- Next install 'Molsoft' nodes. Please uncheck the option "Group items by category" other wise it will not list the nodes.
- After this point everything is setup. You can either go to File/Update KNIME to get the updates manually or setup automatic update.
How to run MolSoft in Knime:
Setting Path to ICM Executable Usually KNIME can figure out where the Molsoft ICM executable is located. In some cases when there are multiple versions of ICM it might return an error as shown below:
Execute failed: Cannot run program "/icm": CreateProcess error=2, The system cannot find the file specified
To fix this:
- File/Preferences
- Select MolSoft and then browse for the location of ICM e.g.C:\Program Files (x86)\Molsoft LLC\ICM-Pro
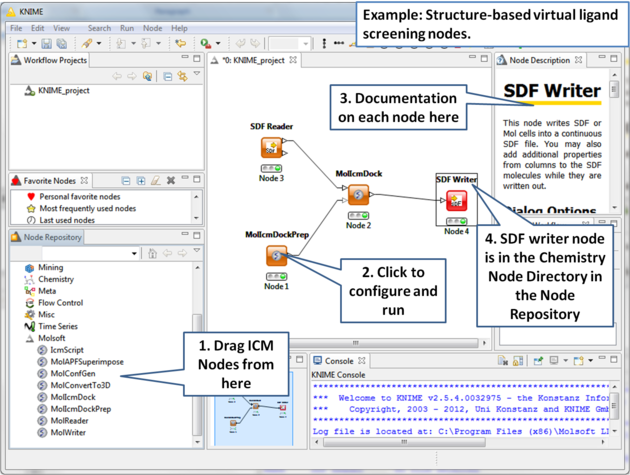
Here is an example for Structure-based virtual ligand screening in KNIME:
| Prev 3DMOLT | Home Up | Next Tutorials |