Newsletter July 2008 |
MolSoft ICM Newsletter July 2008
Welcome to the July 2008 MolSoft ICM Newsletter containing information on:
Fully Interactive 3D Ligand Editor
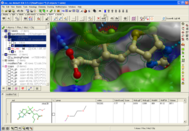 The ligand editor is a powerful tool for the interactive design of new lead compounds in 3D. It allows you to make modifications to the ligand and see the affect of the modification on the ligand binding energy and interaction with the receptor. You can eidt the ligand by adding new functional groups, sampling a large collection of fragments on the fly, edit atoms or bonds, and and then in seconds see the affect of these changes on ligand binding. Clashes with the receptor are clearly flagged and energy contributions of each atom and total bidning energy and binding score are provided. Please see the graphical user interface manual for more information and instructions http://www.molsoft.com/gui/ligand-editor.html
The ligand editor is a powerful tool for the interactive design of new lead compounds in 3D. It allows you to make modifications to the ligand and see the affect of the modification on the ligand binding energy and interaction with the receptor. You can eidt the ligand by adding new functional groups, sampling a large collection of fragments on the fly, edit atoms or bonds, and and then in seconds see the affect of these changes on ligand binding. Clashes with the receptor are clearly flagged and energy contributions of each atom and total bidning energy and binding score are provided. Please see the graphical user interface manual for more information and instructions http://www.molsoft.com/gui/ligand-editor.html
Docking a Markush Library
Generate a Markush library and dock a focused version of it on the fly. See this example, optimizing the roscovitine ligand bound to CDK5 http://www.molsoft.com/gui/markush-docking.html
Export Chemical Spreadsheets to Excel
Export the 2D chemical sketch as well as other column data in a spreadsheet directly to Microsoft Excel. See: http://www.molsoft.com/gui/export-excel.html
Read and Display Google Sketchup 3D Images
 You can now read, render and display any 3D object in ICM. e.g. KMZ and COLLADA format directly from the google sketchup website. We have also added options for changing the texture and material of a 3D object. See http://www.molsoft.com/gui/google-objects.html
You can now read, render and display any 3D object in ICM. e.g. KMZ and COLLADA format directly from the google sketchup website. We have also added options for changing the texture and material of a 3D object. See http://www.molsoft.com/gui/google-objects.html
Lots of New Data Plotting Features From ICM Tables
New plotting features include:
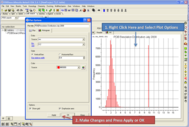
See http://www.molsoft.com/gui/table-plot.html for instructions
Background Images
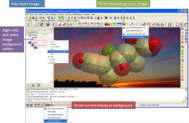 Add any image you want to the background. This is great for making cool images or you can directly compare molecules by making a background image and then making changes to the molecule and see clearly the differences. See http://www.molsoft.com/gui/background.html
Add any image you want to the background. This is great for making cool images or you can directly compare molecules by making a background image and then making changes to the molecule and see clearly the differences. See http://www.molsoft.com/gui/background.html
IUPAC Nomenclature
ICM will now determine the IUPAC nomenclature for any small molecule you sketch or read into ICM. See http://www.molsoft.com/gui/iupac.html
Ribbons and Chain Breaks
Display smooth ribbons and control the display of chain breaks See: http://www.molsoft.com/gui/structure-representation.html#ribbon
Local Chemical Databases
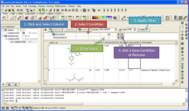 In many cases a compound database (sdf file) is so large that you cannot open it in ICM because you do not have enough memory on your computer. A way around this was to place the database on our product called MolCart but now you can build local chemical databases which you can browse and edit.
In many cases a compound database (sdf file) is so large that you cannot open it in ICM because you do not have enough memory on your computer. A way around this was to place the database on our product called MolCart but now you can build local chemical databases which you can browse and edit.
Some of the features include:
For more information please click here http://www.molsoft.com/gui/local-databases.html
Occlusion Shading
oct08
 The occulusion shading option provides better representation of depth within a cavity. The color of each surface element of a grob (mesh) is changed by mixing its own color with the background depending on the burial of the surface element. See http://www.molsoft.com/gui/mesh-options.html#occlusion-shading
The occulusion shading option provides better representation of depth within a cavity. The color of each surface element of a grob (mesh) is changed by mixing its own color with the background depending on the burial of the surface element. See http://www.molsoft.com/gui/mesh-options.html#occlusion-shading
Protect Objects From Deletion
This option is useful when making molecular documents and you do not want the viewer of the document to delete an object that is contained in the document. See: http://www.molsoft.com/gui/make-molecular-document.html#protect
Wide-Screen Layout
There is now a new layout for users that have a widescreen. Try it by going to File/Preferences/Gui Tab and change GUI.windowLayout
New Command Line Options
Other new command line options are described in the release notes .
ActiveICM - Integrate ICM into PowerPoint or a Web Browser
 ActiveICM will change the way you give presentations or share ICM data with your colleagues. With ActiveICM you can display a series of slides generated in ICM inside PowerPoint or a web browser. The slides are fully interactive which means the user can manipulate the molecules in 3D inside the viewer.
Click here to wownload ActiveICM and see the instructions here http://www.molsoft.com/gui/activeicm.html
ActiveICM will change the way you give presentations or share ICM data with your colleagues. With ActiveICM you can display a series of slides generated in ICM inside PowerPoint or a web browser. The slides are fully interactive which means the user can manipulate the molecules in 3D inside the viewer.
Click here to wownload ActiveICM and see the instructions here http://www.molsoft.com/gui/activeicm.html
The next ICM workshop will be held in La Jolla CA on October 2nd to 3rd. The workshop will be presented by Prof. Ruben Abagyan (Scripps Research Institute, and Founder of MolSoft ) and Dr. Maxim Totrov (Principal Scientist, Molsoft). See http://www.molsoft.com/training.html
Quick Ligand and Peptide Pocket Display
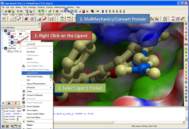 Did you know you can quickly display the surface of a ligand or peptide by pocket by right clicking on the ligand or peptide in the ICM workspace and selecting Ligand Pocket. See http://www.molsoft.com/gui/dsPocket.html
Did you know you can quickly display the surface of a ligand or peptide by pocket by right clicking on the ligand or peptide in the ICM workspace and selecting Ligand Pocket. See http://www.molsoft.com/gui/dsPocket.html