| Prev | ICM User's Guide 7.35 Atomic Property Field Superposition | Next |
[ Pairwise APF Superposition | Flexible APF Superposition to Template | Multiple APF Superposition ]
| Available in the following product(s): ICM-Chemist-Pro | ICM-VLS |
| NOTE: The APF superposition method should be used when there is no common substructure between the chemicals that are being superimposed. If a common substructure is present then the substructure superposition methods described earlier should be used. The APF method will superimpose moieties that have similar 3D pharmacophoric properties. |
The Atomic Property Fields (APF) superposition/alignment method was first reported by Maxim Totrov PhD (Principal Scientist - MolSoft) at the 2007 233rd American Chemical Society National Meeting, Chicago, IL USA and then published here.
APF is a 3D pharmacophoric potential implemented on a grid. APF can be generated from one or multiple ligands and seven properties are assigned from empiric physico-chemical components (hydrogen bond donors, acceptors, Sp2 hybridization, lipophilicity, size, electropositive/negative and charge).Here we describe template APF superposition whereby the APF is generated from a single or multiple template and is then globally optimized with the internal force-field energy of the ligand.
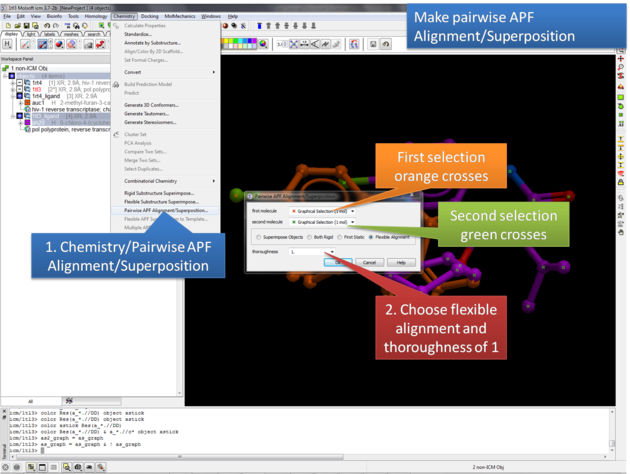
7.35.1 Pairwise APF Alignment/Superposition |
This option allows you to superimipose two chemicals by APF.
- Read in two 3D chemical file or convert a chemical sketch to 3D.
- Select the first chemical by double clicking on it in the ICM workspace. It should be highlighted in blue in the ICM workspace and green crosses in the graphical display. Next, copy this selction to orange (as2_graph)selection to save it - your selection should now have orange crosses.
- Select the second chemical by double clicking on it in the ICM workspace. It should be highlighted in blue in the ICM workspace and green crosses in the graphical display.
- Chemistry/Pairwise APF Superposition/Alignment
- Double check that the first molecule and second molecule selections are correct.
- Choose whether you wish to superimpose both chemical by APF, have both chemicals rigid, first static, or flexible alignment.
- The results will output a new object of the superimposed ligands in the ICM workspace.
7.35.2 Flexible APF Superposition to Template |
To perform Flexible APF Superposition to a template:
- Read a chemical table into ICM containing the compounds you wish to superimpose.
- Display in 3D the template structure you wish to superimpose on. See convert to 3D for instructions on how to generate a 3D template structure.
- Select Chemistry/Flexible APF Superposition to Template and a window as shown below will be displayed.
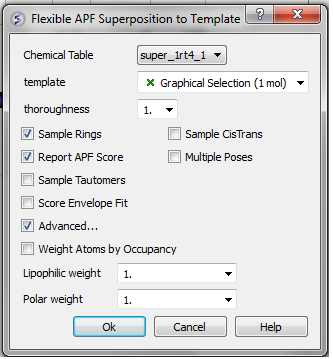
- Use the drop-down arrow to select the chemical table containing the chemicals you wish to superimpose on the template.
- * Select the 3D chemical template by double clicking on it in the ICM workspace. It should be highlighted in blue in the ICM workspace and green crosses in the graphical display.
- Enter an effort value. This represents how long the simulation will run for. A value of 1 has been validated as being a suitable length for this kind of superposition.
- Select whether you want flexible rings to be sampled by checking the appropriate box.
- Select whether you want cis and trans conformations of double bonds to be sampled by checking the appropriate box.
- Select whether you want the superposition to be scored in order to rank solutions by checking the appropriate box. About the APF SCORE - the lower the score the better the fit with the template. No one score or threshold can be stated as being a "good" score it depends on the chemical types being superimposed. One way to tell whether an APF score is good or bad is to compare it to the score obtained by self superimposing the template.
- Select whether you would like to display multiple poses of the superpositions.
- Select whether you would like to sample tautomers.
- Select the Advanced button if you want the superposition to be weighted by occupancy of the atoms by checking the appropriate box. It is often desirable to preferentially superimpose parts of a ligand while ignoring other regions. This can be achieved by setting the occupancy to zero for regions you are not focusing on. See: command line manual http://www.molsoft.com/man/icm-commands.html#set-occupancy . You can also define the weights for lipophilic and polar matches.
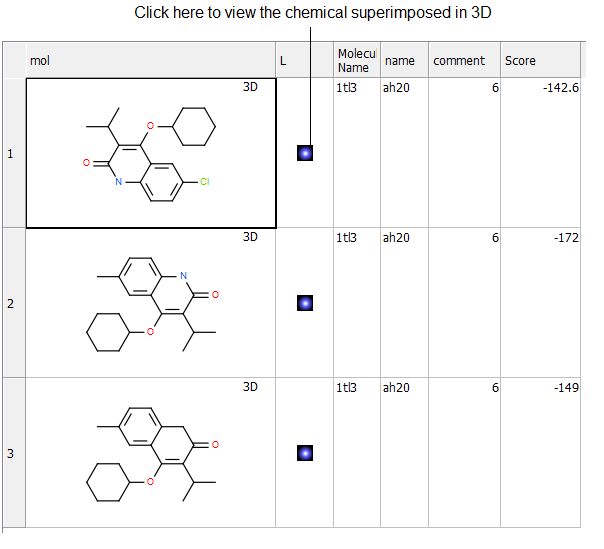
- Click OK and the simulation will run in the background. Once the superposition is complete the molecules will be displayed in the graphical display.
7.35.3 Multiple APF Alignment of Compounds in a Table |
To superimpose multiple chemicals in a chemical table by the APF method:
- Read a chemical table into ICM containing the compounds you wish to superimpose.
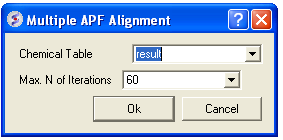
- Select Chemistry/Multiple APF Alignment and a window as shown below will be displayed.
- Use the drop down arrow to select the chemical table.
- Select the number of iterations for the simulation. This represents how long the simulation will run for. A value of 60 has been validated as being a suitable length for this kind of superposition.
- Click OK and the simulation will run in the background. Once the superposition is complete the molecules will be displayed in the graphical display.
The 3D coordinates can be extracted to a chemical spreadsheet as described here.
| Prev Flexible Substructure Superposition | Home Up | Next APF Tools |