Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 5.8 Scan Pockets by 3D Pharmacophore | Next |
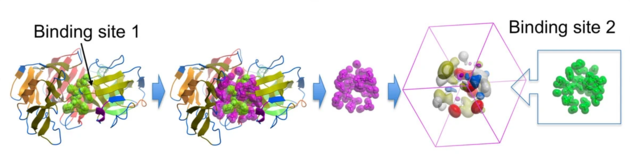
This feature is described in this publication by Max Totrov BMC Bioinformatics 2011.
This binding site comparison algorithm utilizes optimal superposition of continuous pharmacophoric property distributions, this method offers high sensitivity in identifying both distantly homologous and convergent binding sites, demonstrating reliable superposition quality across multiple examples and enabling the effective clustering of ligand binding pockets in PDB.
To run:
- First read in your query protein structure.
- Convert your PDB structure into an ICM object.
- Go to Tools/Scan Pockets
- Either select the ligand and choose the tab "Around Selected Ligands" or make a selection of the pocket and choose the tab "Pocket Atoms" or use ICM PocketFinder and choose the option "Pockets Atom Tags".
- ICM comes with a small compact database to search (pcktmSites.icb.gz) but for a full search use the link in the dialog window to download the full database to search pcktmSitesFull.molt
A results table called SiteHits will be displayed ranking the top scoring PDBs that match the query pocket. The lower the Score value the better the match. Double click on the row and it will superimpose the matched structure onto the query.
| Prev 3D Similarity | Home Up | Next Convert to ICM Object |