ICM-Bio |
| Watch ICM-Bio in Action |
The ICM-Bio provides the environment for manipulation of sequences, alignments, profiles and databases both interactively and in a batch mode. It also provides direct connectivity with 3D models and a fully interactive alignment editor and annotator.This package also contains all the features in ICM-Browser-Pro.
Accurate pairwise alignments and their significance: ICM includes two high-quality global pairwise alignment algorithms with zero-end penalties (ZEGA). double-affinity gap penalties and dual insertions. Also an empirical statistics was derived for many residue substitution matrices and gap penalties to evaluate the probabbility that the two aligned sequences share the same structural fold.†This statistics can also be used locally to evaluate the local reliability of an alignment, or identify the alternative alignments and repeats in probability weighted dot-plot.
Alignment editor: ICM has an easy to use editor for pairwise and multiple alignments. ICM alignment editor is robust and always protects the integrity of your alignment by protecting you from making unintended changes in the alignment. You can edit the alignment, shade, box, and annotate the alignment. The alignment can be exported as an image or in other alignment formats.
ZEGA-based database searches for distance relatives and structural similarities. The sensitive and rigorous Zega alignment is used for each comparison, and the structural statistics identifies the significant hits.†This search may give you more homologues than a BLAST search! The output may be presented in a linked table form:
Pattern searches, prosite and profile searches, other functions. Other functions include tools for indexing and quering the flat-file sequence databases, annotation transfer functions, sequence-structure connectivity.
Multiple Sequence Alignments and principal component clustering: Accurate pairwise sequence-sequence, sequence-profile and profile ñprofile alignments are used to build multiple sequence alignments with a hierachical, evolutionary tree-guided, alignment procedure.†Better pairwise alignments, neighbor-joining tree construction algorithm and weighted profile construction leads to high-quality alignments. The shell allows to generate them in batch and control all the alignment parameters.
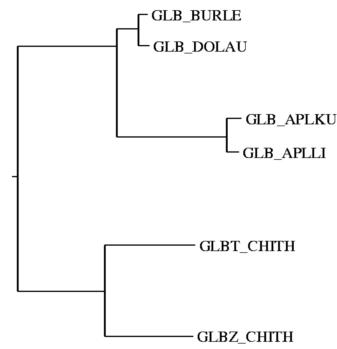
Sequence Structure Alignment: Use an interactive alignment editor for easy alignment and tether editing. Color your selections for easy identification. Relationships between sequences can be shown as evolutionary trees from the specified sequence alignment using the neighbor-joining method (Saitou and Nei, 1987), as 2D distribution of sequences using the two main principal axes, and as 3D distribution. This can be analyzed in stereo using controls of molecular graphics.†Create a PostScript images of your results.
Sequence-Structure Alignment for Homology Modeling: Both the secondary structure and the template 3D structure are used for sequence threading.†Search your sequence interactively or in batch through any database and generate a list of possible homologues which are sorted and evaluated by probability of structural significance.†Use this alignment in homology modeling.
| Sequence Analysis |
| Read in sequences and alignment in many different formats including FASTA. |
| Direct link to the UniProt sequence database. |
| Direct link to PFam. |
| Calculate sequence residue content a table and graph of residue frwequencies will be displayed. |
| Predict sequence secondary structure. |
| Six frame translation. |
| Set sequence type e.g Protein or DNA. |
| Fully interactive sequence editor. |
| Save sequence as an image. |
| Save a sequence in a variety of formats. |
| Sequence Alignments |
| Align two sequences. |
| Seven alignment comparison matrices to choose from. |
| A variety of alignment methods available including ZEGA, H-align, Needleman-Wunsch, Smith-Waterman and Wilbur-Lipman. |
| Adjust parameters such as Gap Open, Gap Extension. |
| Perform sequence to structure alignment. |
| Align DNA to Protein. |
| Multiple Sequence Alignments |
| Dynamic and feature-rich multiple sequence alignment editor. |
| Easy drag and drop alignment construction. |
| Link the alignment to 3D protein structures in the graphical display. e.g. select a region in the alignment and this selection will be propogated onto the protein structure. |
| Extract a sub alignment. |
| Cut a vertical alignment block. |
| Reorder sequences in an alignment. |
| Extract unique sequences. |
| Hide/Show gaps in the alignment. |
| Compress gaps in the alignment. |
| Search in an alignment. |
| Save and export a high quality image of the alignment. |
| Copy the alignment to clipboard. |
| Build a residue number table of the alignment. |
| Build a residue label table of the alignnent. |
| Cut a selected block of an alignment. |
| Make a sequence QSAR table. |
| Copy and paste selected regions of an alignment. |
| Annotate regions of an alignment. |
| Shade regions of an alignment. |
| Change the font color in an alignment. |
| Set fonts to bold. |
| Draw a box around a region of an alignment. |
| Add a title to an alignment. |
| Display the consensus sequence of the alignment. |
| Display the sequence profile at each point of the alignment. |
| Display sequence ruler. |
| Display sequence tree. |
| Display the sequences in the alignment as unaligned. |
| View differences in the alignment. |
| Color the alignment by a variety of built-in and user-defined color schemes. For example you can adjust the consensus strength and color or color by hydrophobicity. |
| Make complex selections in the alignment. This is helpful when the alignment is linked to a 3D structure. |
| Display sequence secondary structure. |
| Sequence Search |
| ICM-Pro provides a fast tool to search any Blast-formatted database with a query sequence and generate an alignment on the fly. |
| Search by sequence similarity. |
| Search by sequence homology. |