Google Search: Keyword Search:
| Prev | ICM User's Guide 7.12 Annotate By Substructure | Next |
| Available in the following product(s): ICM-Chemist | ICM-Chemist-Pro | ICM-VLS |
This feature allows you to annotate a chemical spreadsheet according to functional group. It also allows you to flag substructures which may have poor ADME properties.
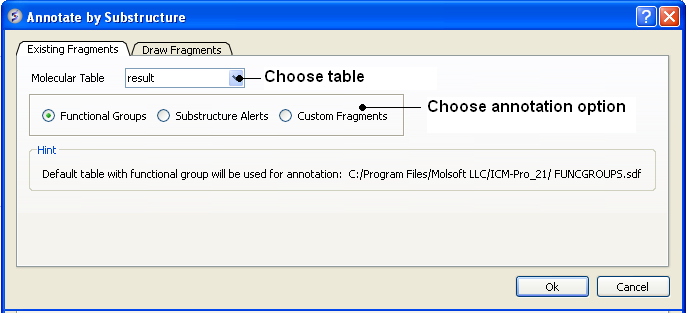
First read in a chemical spreadsheet or sdf file you wish to annotate. To do this:
- Information on how to read into ICM chemical spreadsheets and structure can be found here.
To annotate functional groups:
- Chemistry/Annotate by Substructure.
- Enter the name of the Molecular Table (Chemical Spreadsheet) or use the drop down button to locate it.
- Check the Functional Groups option.
- The functional groups will be listed in a new column in your chemical spreadsheet called funcgroup. The default table with functional group will be used for annotation called FUNCGROUPS.sdf in ICMHOME.
To annotate potentially poor ADME groups (Substructure Alert).
- Chemistry/Annotate by Substructure.
- Enter the name of the Molecular Table (Chemical Spreadsheet) or use the drop down button to locate it.
- Check the Substructure Alerts option.
- The alerts will be listed in a new column of your chemical spreadsheet called alerts. The default table with substructure alerts will be used for annotation called CHEMFILTER.sdf
To annotate by custom fragments
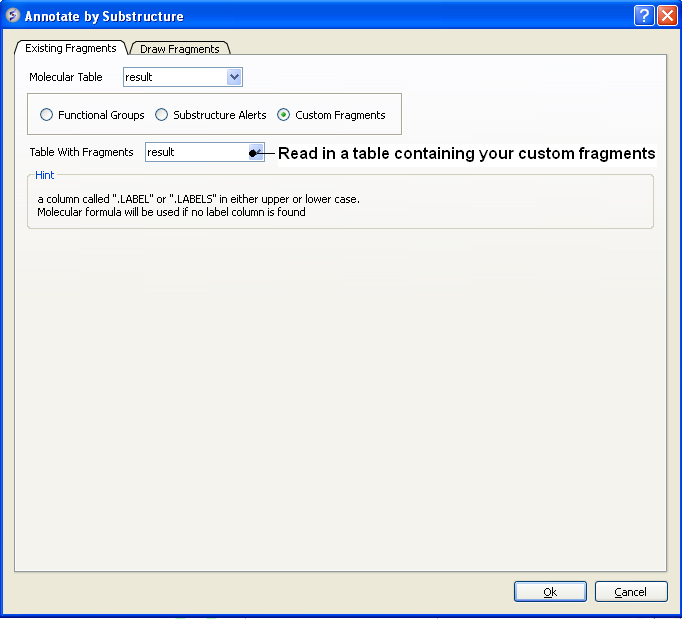
- Read into ICM a table with annotated fragments. The fragments should be labeled in a column called ".LABELS" or ".LABEL" in either upper or lower case. If not "LABELS" column is found the molecular formula will be used instead.
- Chemistry/Annotate by Substructure.
- Check the Custom Fragments button
To draw a fragment and annotate.
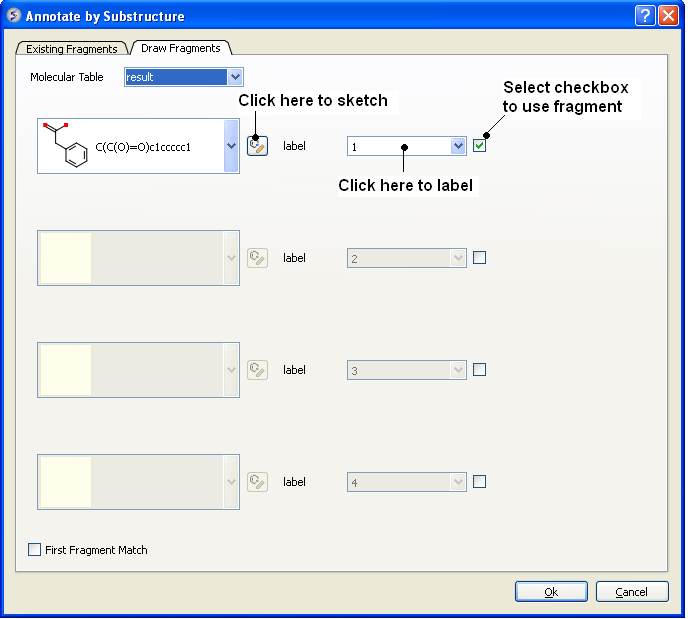
- Click on the Draw Fragments tab.
- Enter the SMILES string of the fragment or click on the ligand editor button in the dialog window to sketch the fragment.
- Enter a label name.
- Click the check box next to the label name.
- Click OK
| Prev Standardize Table | Home Up | Next Align/Color by 2D Scaffold |