Google Search: Keyword Search:
| Prev | ICM User's Guide 7.1 Read Chemical Structures | Next |
[ Extract from PDB | Large SDF Files | Extract 3D ]
| Available in the following product(s): ICM-Chemist | ICM-Chemist-Pro | ICM-VLS |
| Video
Chemical structures can be read into ICM from MOL/MOL2, SMILES, and SDF files or you can construct your own structures by drawing them in the ICM Molecular Editor. Chemicals can also be searched and downloaded via the Search Tab using the PDB Chemical Substructure Search, search by PDB Ligand Code, or by searching DrugBank, ChEMBL, SureChEMBL or PubChem.
To read a chemical format file:
- Select File/Open.
- Select the chemical structure file you wish to open: MOL, MOL2 or SDF.
- Once selected the file will be displayed as a chemical table (See ICM molecular tables section).
If you know the chemical smiles string for the compound you can build the 2D molecule by:
- Select File/New.
- Click the Compound tab at the top of the window.
- Enter a name for the compound.
- Type in the Smiles String in the Smiles String data entry box. Remember to delete the previous string.
- Check the boxes Display Molecule Delete Other Objects according to your preference.
- Click the OK button.
Smiles can be read from a text file into a chemical table by:
- File/Open and select Files of type: Smiles format
- Then you can convert SMILES to 2D sketch using Chemistry/Convert.
Smiles can be read directly into the ICM Molecular Editor:
- Open the ICM Molecular Editor window.
- Select Edit/Add Smiles
Multi mol2 3D files can be read by:
- File/Open and select Files of type: mol2 format.
Chemicals can be read directly from ChEMBL, SureCHEMBL, PubChem, and DrugBank
- Click on the search tab and find the database you wish to search in the drop down dialog box.
7.1.1 Extract 2D structure from PDB Chemical |
To extract a 2D chemical representation from a PDB file into a chemical spreadsheet.
- Read the PDB file.
- Right click on the ligand and choose Extract Ligand.
- Choose whether you want to extract the 3D coordinates.
- Choose whether you want to append to an existing table (chemical spreadsheet).
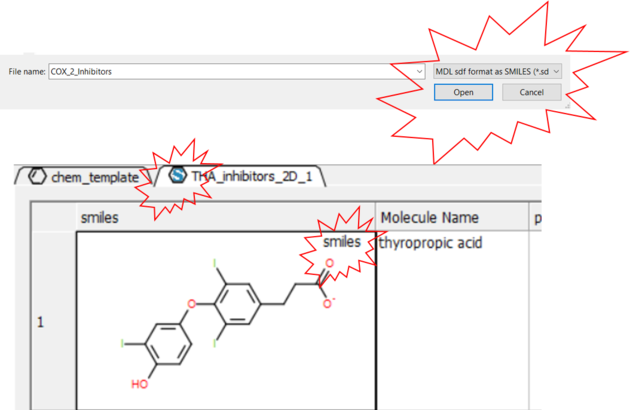
7.1.2 Read Large SDF Files |
The ability to read large SDF files (e.g. >100K chemicals) depends on the memory of your computer. A work around for this is available in ICM.
- File/Open and in the Filter Section choose "MDL sdf format as SMILES"
- The table header will contain an "S" symbol representing that the table contains a SMILES column rather than mol. Each cell will also be labeled "Smiles".
7.1.3 Extract 3D coordinates from 3D Chemical. |
To extract the 3D coordinates of a chemical into a chemical spreadsheet e.g. for 3D QSAR or after chemical superposition.
- Select the 3D chemicals in the ICM workspace. One or more can be selected by clicking holding the CTRL key and clicking. A range of chemicals can be selected by holding the keyboard tab and clicking.
- Right click on any of the selected chemicals in the ICM workspace and choose Extract Ligand.
- Select "keep 3D coordinates.
- Append to an existing table if needed.
| Prev Cheminformatics | Home Up | Next Save |