Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 12.10 Ligand AIDE: AI Design Evolution | Next |
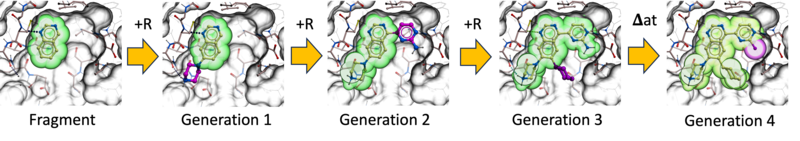
Ligand AIDE is a de novo ligand generation workflow based on Artificial Intelligent Design Evolution. It uses an iterative evolutionary strategy to grow and optimize ligands directly in the context of a protein binding site.
The process begins with a population of docked small fragments. From this starting point, new designs are generated by adding or replacing R-groups using the groupGen neural network, as well as by performing atom-level substitutions using the Atom Predictor. Each newly generated cohort of compounds is then re-docked into the binding site.
Designs are refined through multiple evolutionary cycles, typically three to five iterations. At each step, a Darwinian selection process removes poor designs based on several criteria, including RTCNN scores, physics-based binding scores with penalty terms, drug-like property filters, and synthesizability.
Through successive generations, fragments evolve into more complete and optimized ligands. This iterative grow-and-select strategy enables efficient exploration of chemical space while maintaining binding quality, drug-like behavior, and practical synthetic feasibility.
To run Ligand AIDE:
- Setup the Docking project.
- Docking -> De Novo Tools -> AI Ligand Generator
- Enter the name of the Docking project.
- Enter the Number of Generations. This option controls the size of the result ligand. Default (4) is generally for mid-size ligands. For large pockets the value can be increased.
- Enter Ligand Population number. This option defines how many ligands are retained and evolved in each generation, balancing chemical diversity against runtime.
- Enter the Min Synthesizability cutoff. Sets the minimum synthesizability score required for ligands to survive selection, enforcing practical synthetic feasibility.
Interpreting Ligand Aide Output Columns
This section explains several columns in the Ligand Aide results table and how they should be interpreted.
Generation
The generation column reports the number of growth iterations used to build the ligand.
The goal of Ligand Aide is to generate a ligand that fits into the binding pocket. On each iteration, the algorithm attempts to grow a chemical group into available space within the pocket.
Four generations is typically an average number of iterations required to produce a ligand that fits an average-sized pocket. The final number of generations depends on several factors:
- Size and position of the initial fragment
- Size and shape of the pocket
The value in the generation column is primarily informational and should not be used to prioritize results.
FreeStrain
The FreeStrain value estimates the conformational strain of the ligand.
It is calculated using a neural network based torsion profile method described here:
https://pubmed.ncbi.nlm.nih.gov/36456533/
Additional details are available at:
https://www.molsoft.com/gui/torsion-free-strain.html#gsc.tab=0
SumScore
The SumScore is a weighted combination of multiple scoring terms and is not simply the sum of Score and RTCNN.
It is calculated as:
VlsScore * (1 - wRTCNN) + RTCNN * wRTCNN + FreeStrain
where:
wRTCNN = 0.33
Synthesizability Score
A synthesizability cutoff is applied before Ligand Aide is run. If needed, a synthesizability score can be calculated afterward.
This can be done using:
Chemistry -> Properties -> MolSynth
The score is not currently included in the default results table but is straightforward to compute after the run.
| Prev chimeric | Home Up | Next Autofit |