Google Search: Keyword Search:
| Prev | ICM User's Guide 11.16 Covalent Docking in the Ligand Editor | Next |
| Available in the following product(s): ICM-Pro | ICM-VLS | ICM-Chemist-Pro |
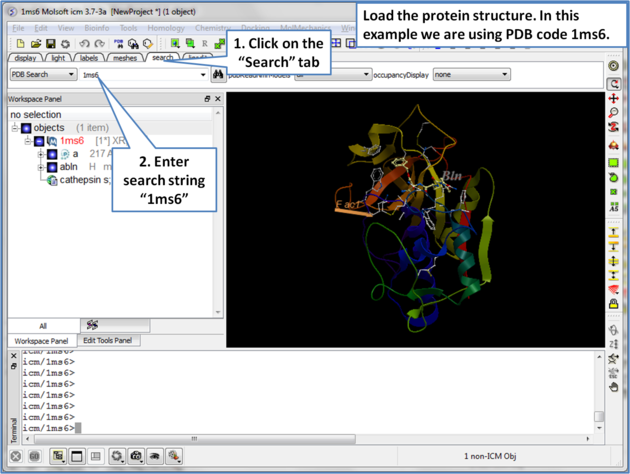 |
| Step 1: Load the protein structure via the Search tab. In this example we will use PDB structure 1ms6. |
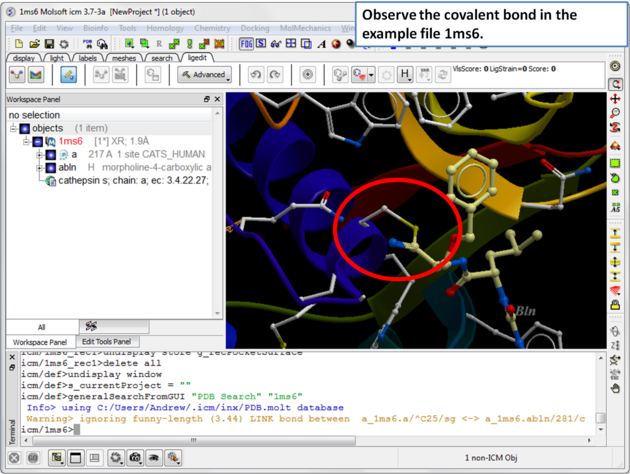 |
| Step 2: The structure 1ms6 has a covalent bond between the ligand "abln" and the receptor residue C25. |
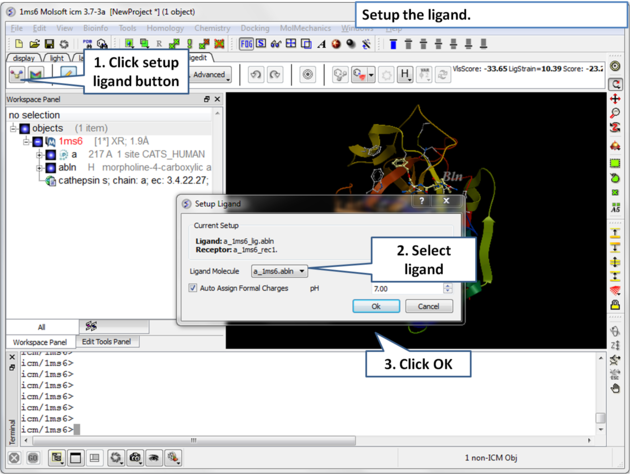 |
| Step 3: Choose the ligand "abln" and setup the ligand in the regular way. |
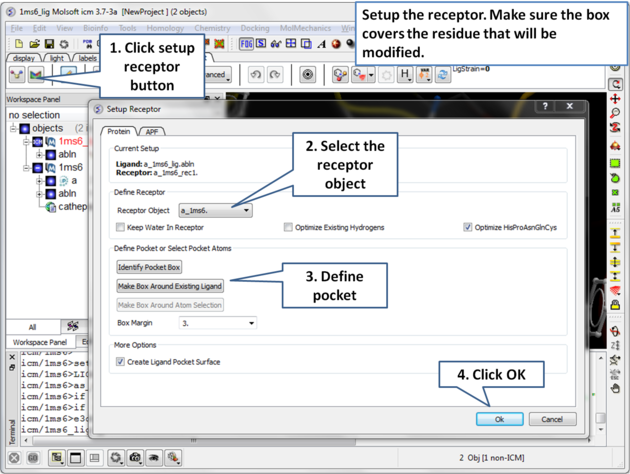 |
| Step 4: Setup the receptor and make the maps. It is important to make sure the box encompasses the modified residue C25. |
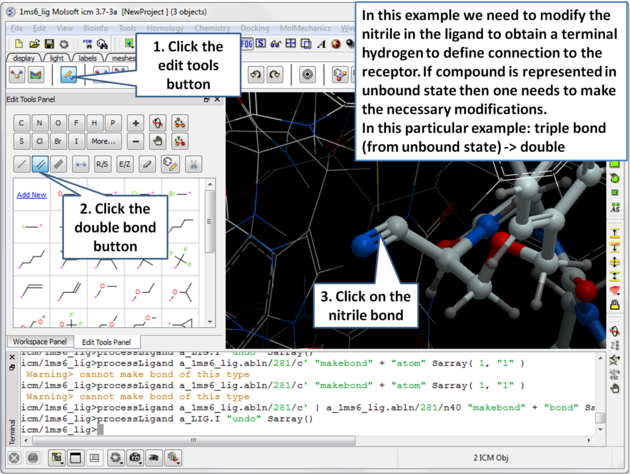 |
| Step 5: Modify the nitrile in the ligand to obtain a terminal hydrogen to define the connection to the receptor. |
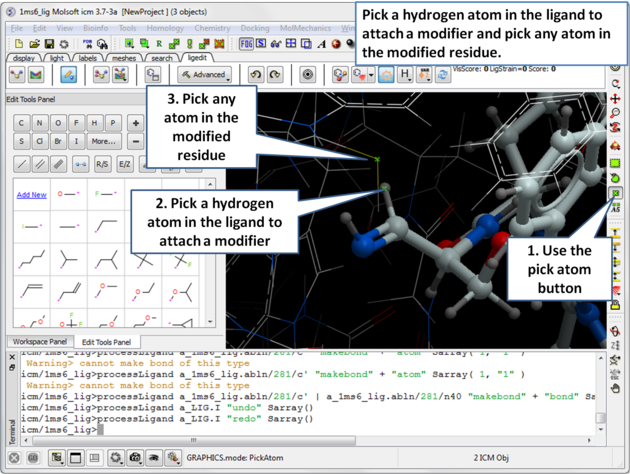 |
| Step 6: Pick the hydrogen atom in the ligand where the modifier will be attached and then pick any atom in the modified residue. |
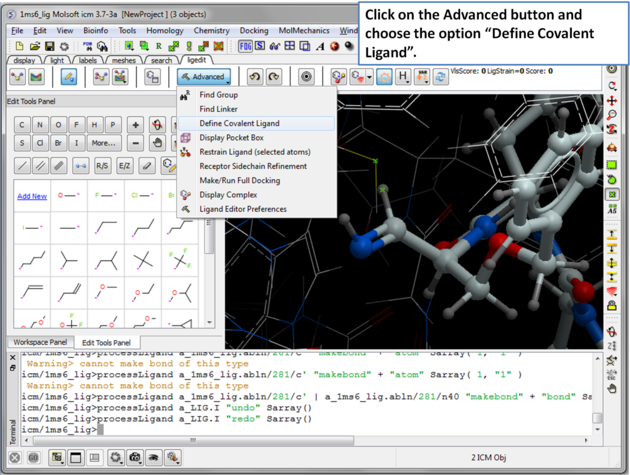 |
| Step 7: Click on the Advanced button and choose the option "Define Covalent Ligand". |
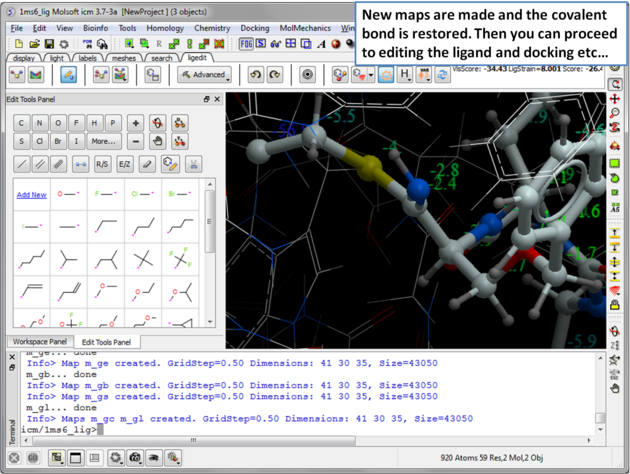 |
| Step 8: New maps will be made and the covalent bond is restored. Then you can re-dock, minimize and edit the ligand in the usual way inside the ligand editor. |
| Prev Multiple Receptor Docking | Home Up | Next Dock to APF |