Google Search: Keyword Search:
| Prev | ICM User's Guide 15.2 Creating Fully Interactive Slides for PowerPoint and the Web Tutorial | Next |
Introduction
A slide enables you you to conveniently store a large number of different 3D visualization properties (e.g. color, viewpoint, representation etc...) along with text and window layout. ActiveICM enables you to view and display ICM graphical slides and animations interactively inside Windows Microsoft PowerPoint and web browsers such as Internet Exporler and Mozilla Firefox.In this example we are going to look at the effect of an agonist binding to the Beta-2-Adrenergic GPCR (B2AR) and prepare a molecular document. A prepared ICM file containing three B2AR GPCRs superimposed can be downloaded here https://molsoft.com/addons/ftp/workshop (1. cut and paste the address into a web browser. 2. Right click on GPCR_slides.icb and choose "Save Link As". 3. Open the .icb in ICM File/Open). The file contains pdb files 2RH1 (R inactive state), 3P0G (R* active state, with G-alpha mimic), and 3SN6 (R*G G-protein signalling state). The agonist ligand causes Helix 5 to shift inwards and Helix 6 to swing outwards. This agonist binding poses were predicted using ICM a year before the crystal structures were solved, you can read about this here and a good review of the structure and function of GPCRs here.
Tutorial
In this tutorial we will build a series of slides and then annotate them using HTML to create hyperlinks to the slides. The resulting molecular document can be exported in movie format or in fully interactive 3D files.
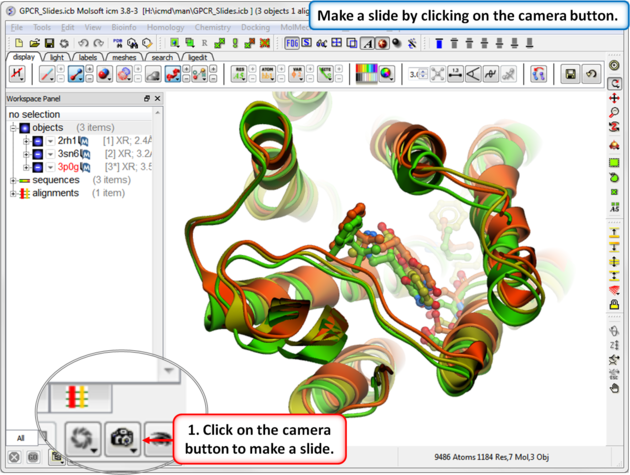 |
| Step 1: Download the example file here https://molsoft.com/addons/ftp/workshop (1. cut and paste the address into a web browser. 2. Right click on GPCR_slides.icb and choose "Save Link As". 3. Open the .icb in ICM File/Open). Make a slide by clicking on the camera button at the bottom of the GUI. |
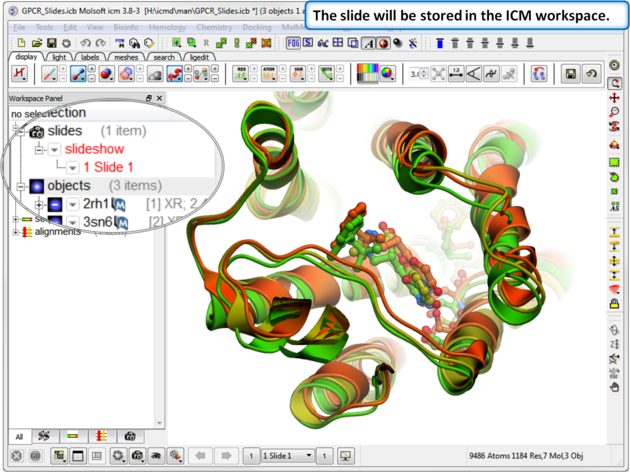 |
| Step 2: The slide is stored in the ICM Workspace panel (left hand side of the GUI). You can make changes to the 3D display and return to the display in the slide by clicking on the slide in the ICM Workspace. |
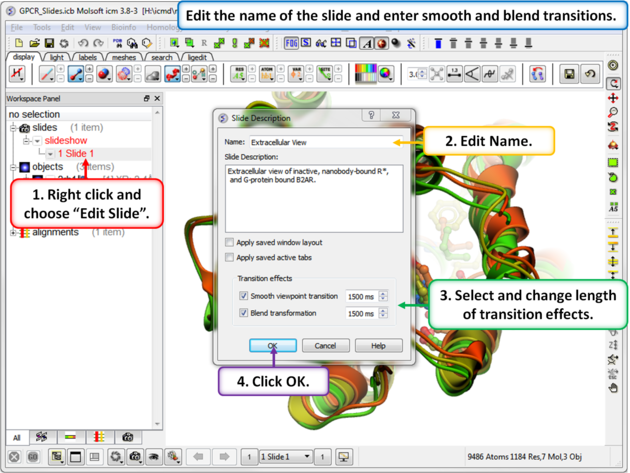 |
| Step 3: Properties of the slide can be edited by right clicking on the slide in the ICM workspace. |
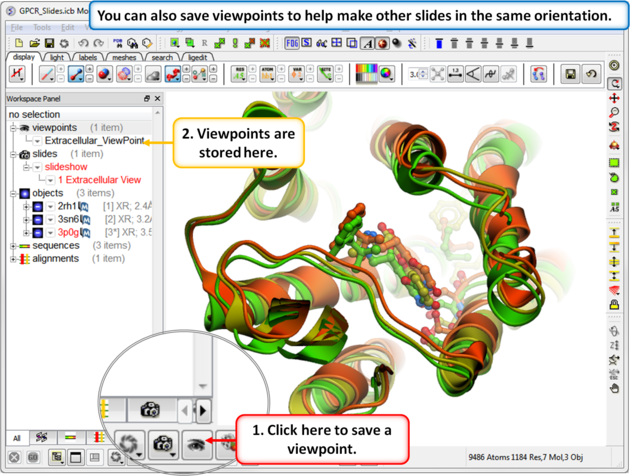 |
| Step 4: Saving a viewpoint is useful if you are making many slides from the same viewing position. |
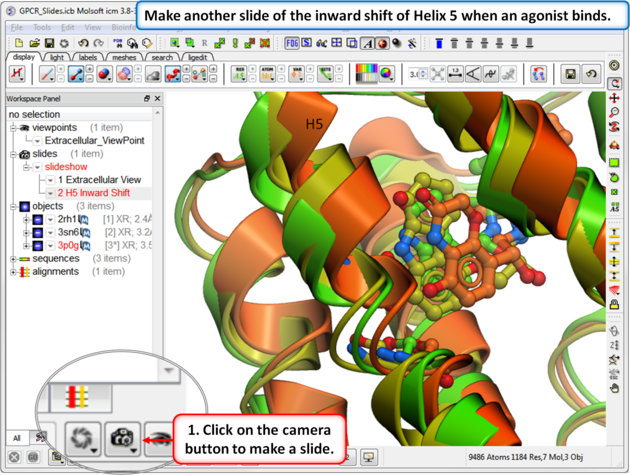 |
| Step 5: In this slide we show that Helix 5 makes an inward shift when an agonist binds. |
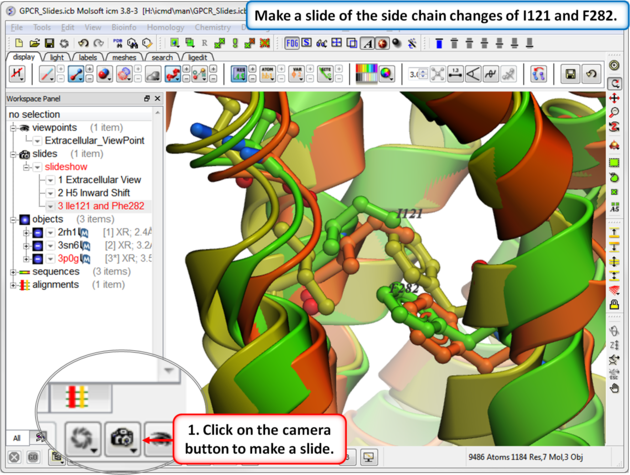 |
| Step 6: In this slide we show that upon binding an agonist Ile121 switches its rotameric state and Phe 282 moves. |
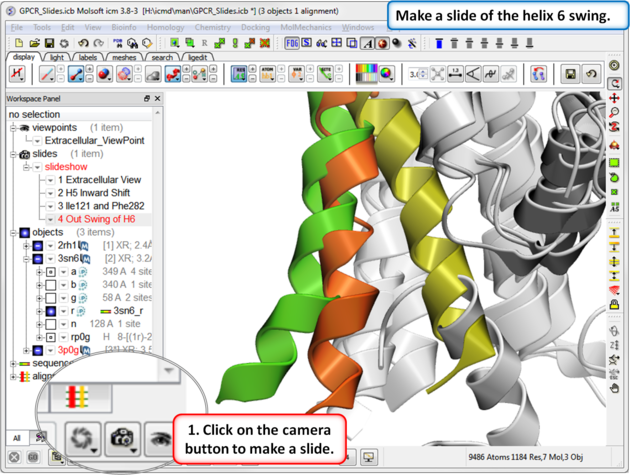 |
| Step 7: In this slide we show that agonist binding causes Helix 6 to swing out. |
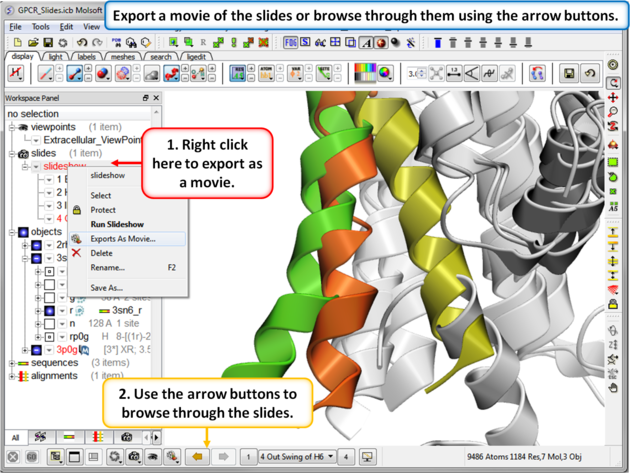 |
| Step 8: You can export the slides as a movie or you can browse through them using the arrow buttons. |
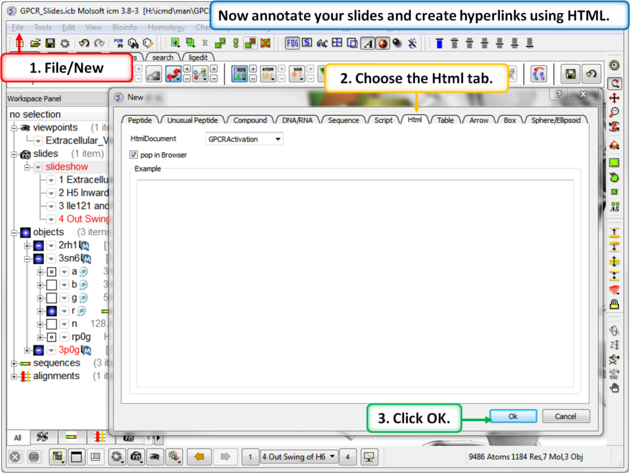 |
| Step 9: To build a molecular document you can annotate the slides using HTML. |
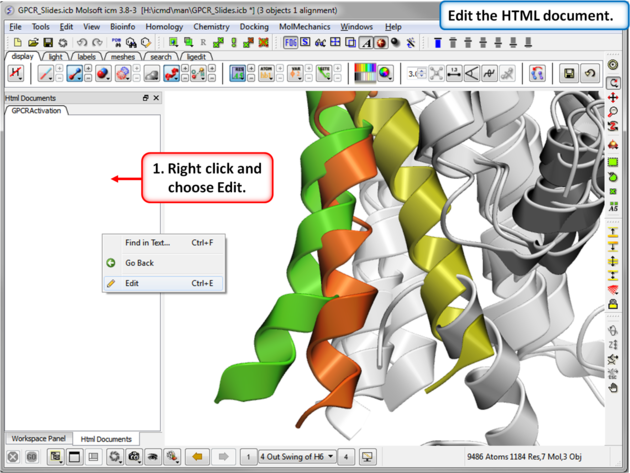 |
| Step 10: Edit the document by right clicking on the HTML panel and choosing Edit. |
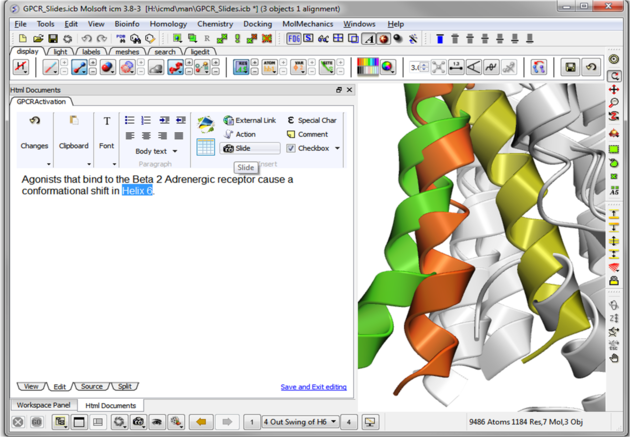 |
| Step 11: Hyperlinks can be made to the slide. Other functions such as adding check boxes, ICM scripts and external links are also available. |
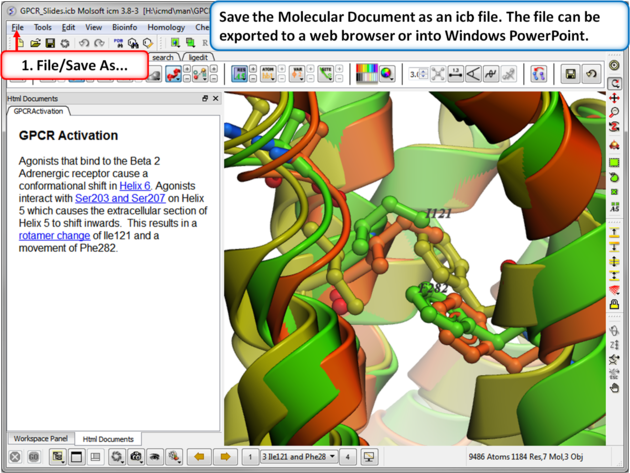 |
| Step 12: Save the molecular document in icb format and view in ICM-Pro or ICM-Browser. You can also export to a web browser or view the fully interactive slides in Windows Powerpoint using a plugin called ActiveICM. |
| Prev 2D and 3D Labels | Home Up | Next Protein Structure Tutorials |