Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 16.18 AI Decoration (groupGen) | Next |
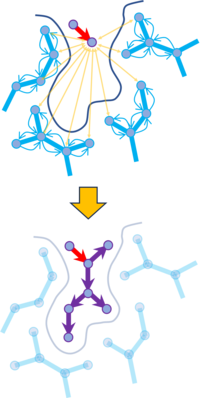
AI decoration (groupGen) is a structure-aware generative neural network developed for R-group optimization in lead refinement workflows. The method generates chemically valid substituents at a user-defined attachment point by explicitly modeling the three-dimensional protein environment surrounding that position. The input consists of a fixed ligand core, a defined attachment atom, and the corresponding receptor structure, allowing groupGen to focus generation on a localized region of the binding site.
The encoder component is implemented as a 3D graph convolutional neural network that samples atomic features within a defined radius around the attachment point. Protein and ligand atoms are represented using geometric, electrostatic, and hydrophobic descriptors, which are aggregated into a latent pocket embedding. This embedding captures steric constraints and interaction preferences that guide substituent selection.
The decoder is based on MolSoft's Sprouts chemical graph generation architecture and is pre-trained as a variational autoencoder on large collections of chemically valid R-groups. During inference, the decoder is conditioned on the pocket embedding and constructs substituent graphs that are compatible with the attachment point and optimized for favorable interactions within the binding pocket.
Generated analogs are passed directly into the validation pipeline. CombiRidge performs anchored docking to preserve the core ligand pose while optimizing substituent conformations. Resulting poses are evaluated using RTCNN and VLS scoring functions to assess binding quality, followed by a synthesizability score to deprioritize chemically impractical structures. The final output is a ranked list of R-group substitutions optimized for binding, geometry, and synthetic feasibility.
To run the AI Decoration in the ligand editor:
- Setup the ligand and receptor in the ligedit tab.
- Click on the Advanced menu button and choose AI Decoration.
- Number of AI steps controls the number of iterative generation steps used by the groupGen decoder to construct an R-group. Higher values allow larger and more diverse substituents to be generated, while lower values bias the model toward smaller, simpler modifications and faster generation.
- You can choose to increase the number of compounds to rescore.
When completed you will see a new table called NEWGEN_AI. The new designs are ranked by Score Combined which is the sum of the physics based score and the RTCNN score. You can double click on a row to load the new design into the receptor to edit it or use the 'L' column button to toggle its display it. The table also reports the synthesizability score (>0.5 is considered synthetically feasible) and the Strain.
| Prev Dock to APF | Home Up | Next Save Docked Complex |