Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 12.3 Run the Docking Simulation | Next |
[ On-screen Demo Dock | Batch Docking | Run Docking Batch | Dock Chemical Table ]
Once the receptor and maps have been correctly set up then the docking procedure can begin.
There are three possible options to choose when On-screen Demo Dock, Setup Batch Ligand, or Dock Chemical Table docking. (Please note some of the options may be limited for users without ICM-VLS)
- On-screen Demo Dock is ideal when you have a single ligand and simply want to re-dock it to the receptor. For anything more advanced, such as multiple ligands or larger workflows, use Batch Docking or Dock Chemical Table (see below). It can also be used to view the ligand docking to the pocket which is useful for teaching or demonstration purposes. In ICM versions 3.8-5 and older this option was called Interactive Docking.
- BATCH docking will run in the background and is an ideal choice for screening a database of compounds e.g. vls.
- Dock Chemical Table will allows you to easily dock a chemical spreadsheet / sdf file that is loaded into ICM.
12.3.1 On-screen Demo Dock |
[ On-screen Demo Dock: Mol Table Ligand | On-screen Demo Dock: Loaded Ligand ]
This option is useful for visualizing ligand binding, making it ideal for teaching or demonstrations. For docking research projects use Setup Batch Ligand, or Dock Chemical Table docking. Use On-screen Demo Dock docking to dock one ligand at a time in the foreground. It is ideal to use this option for small-scale docking and to view the flexible ligand sampling the pocket.
- Click on the menu Docking/On-screen Demo Dock
Choose either Mol Table Ligand or Loaded Ligand
12.3.1.1 On-screen Demo Dock: Mol Table Ligand |
If you have a chemical table already loaded into ICM you can use this option to dock chemicals in it. You can read mol/mol2 or sdf files into ICM by using File/Open. They will be displayed in a table.
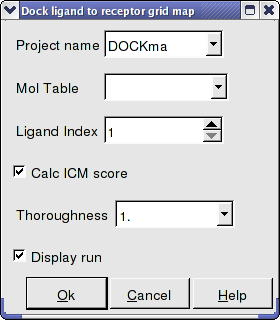
- Enter the Docking Project Name
- Use the drop down arrow to find the table of ligands you wish to dock.
- Enter the number of the ligand in the table you wish to dock. For example, if the ligand is in row 6 enter 6.
- If you have ICM-VLS you can retrieve an ICM docking score for the docked ligand.
- 'Effort' or thoroughness represents the length of the simulation. Generally 1 is a reasonable value for buried hydrophobic pockets. If you are docking to solvent exposed pockets or pockets containing metal ions you may wish to increase this parameter.
- Display run will display the ligand sampling the energy in the ligand binding pocket. Although this is fun to watch this significantly slows down the docking operation.
12.3.1.2 On-screen Demo Dock: Loaded Ligand |
If you have a ligand as an ICM object you can use this option.
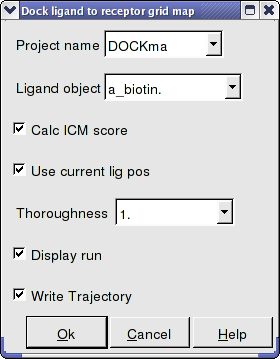
- Enter the Docking Project Name.
- Use the drop down arrow to find the ligand.
- If you have ICM-VLS you can retrieve an ICM docking score for the docked ligand.
- If the ligand is already located in the pocket you can use this option. However by default the ligand will start sampling in the center of the pocket so this option does not need to be used.
- Thoroughness represents the length of the simulation. Generally 1 is a reasonable value for buried hydrophobic pockets. If you are docking to solvent exposed pockets or pockets containing metal ions you may wish to increase this slightly.
- Display run will display the ligand sampling the energy in the ligand binding pocket. Although this is fun to watch this significantly slows down the docking operation.
- You can write the docking simulation to a trajectory file. Please see the command language manual for more information on this.
12.3.2 Batch Docking |
[ From Loaded ICM Object | From File | From Mol/Mol2 | From Indexed Database | From MolCart Database ]
Batch Docking is used for running docking jobs in the background. It is ideal for large-scale docking and virtual ligand screening jobs.
- Use the menu option Docking/Batch Ligand Setup to setup the docking run.
- Once setup use the menu option Docking/Run Docking Batch to run the batch docking.
12.3.2.1 Batch Docking: From Loaded ICM Object |
From Loaded ICM Object Your ligand needs to be an ICM object and loaded into ICM (File/Open). Your object will be displayed in the ICM Workspace.
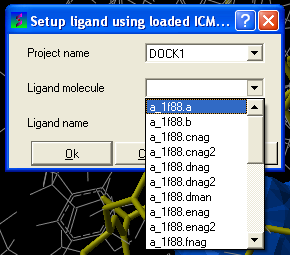
- Enter the name of the docking project.
- Ligand Molecule Use the drop down button to find the loaded ligand or enter the ICM selection language.
- Ligand Name A docked ligand will be given a name. Change this if you wish or use the default name.
- Once setup use the menu option Docking/Run Docking Batch to run the batch docking.
12.3.2.2 Batch Docking: From File |
From File: ICM If your ligand (s) is saved and converted to an ICM object but is not loaded into ICM then you can use this option.
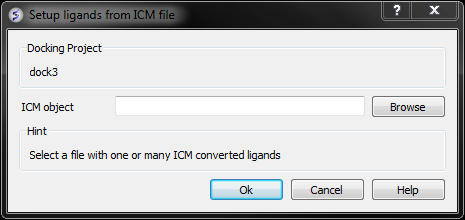
- Browse for the ICM Object.
- Click OK
12.3.2.3 Batch Docking: From Mol/Mol2 |
From File:MOL/MOL2
If your ligand is a MOL or MOL2 file then
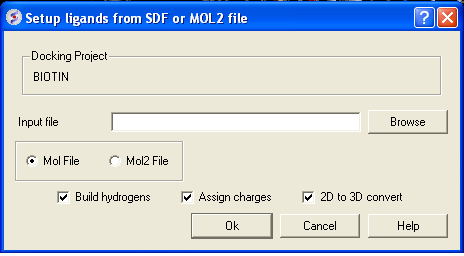
- Browse for your MOL/MOL2 file.
- Select whether your ligand is in MOL or MOL2 format.
- If you wish hydrogens to be added to your compound or charges to be assigned then click on the appropriate boxes in the display panel.
- Click OK
12.3.2.4 Batch Docking: From Indexed Database |
From Indexed Database - only available with ICM-VLS
In most cases the ligand input file will be an SDF or MOL2 file. These files need to be indexed by ICM before they can be used in VLS runs (see next section of this manual). The index is used to allow fast access to an arbitrary molecular record in a large file such as an SDF file which in some cases contains over one million compounds.
To index an sdf file:
- Click on the menu Docking/Tools/Index Mol/Mol2 File/Database to generate the index. The following data entry box will be displayed.
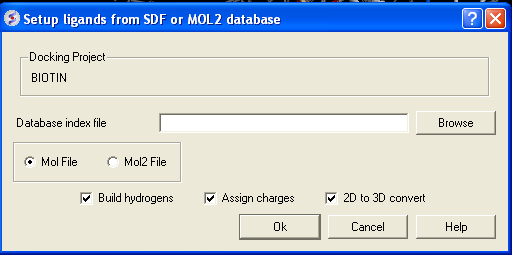
- Enter the name of your Mol/Mol2 file and enter the name you wish to call your index file.
- Select whether your file is in Mol or Mol2 format.
- Browse for your Index file.
- Select whether your ligand is in MOL or MOL2 format.
- If you wish hydrogens to be added to your compound or charges to be assigned then click on the appropriate boxes in the display panel.
- Click OK
12.3.2.5 Batch Docking: From MolCart Database |
From MolCart - only available with ICM-VLS
| NOTE A separate license is required for MolCart |
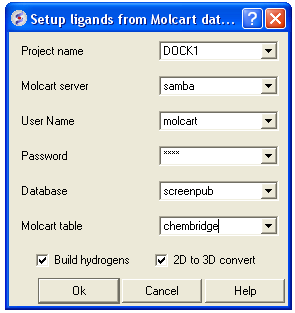
- Enter docking project name.
- Enter the MolCart server
- Enter your username
- Enter Password
- Enter the Database Name
- Enter the name of the MolCart table within the Database
- Select whether you would like to build hydrogens or convert the compounds from 2D to 3D.
12.3.3 Run Docking Batch |
IMPORTANT Once you have setup the docking you need to set it running using the menu option Docking/Run Docking Batch
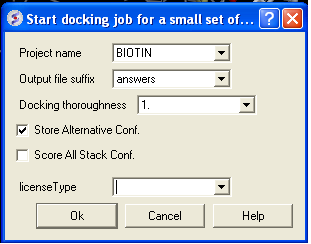
- Docking/ Run Docking Batch
- Enter the Docking Project Name
- Batch docking will generate an output file - enter the name you wish to call this file here.
- Thoroughness represents the length of the simulation. Generally 1 is a reasonable value for buried hydrophobic pockets. If you are docking to solvent exposed pockets or pockets containing metal ions you may wish to increase this slightly.
- Selecting Store Alternative Conf will allow you to look at all conformations in the energy stack.
- Score All Stack Conf. will allow you to determine an ICM docking Score for all members of the stack - this will slow the docking down.
- License type: For nearly all license (e.g. standard ICM-VLS licenses) types you need to leave this entry blank. If you have a -vlscluster license or a molvls license (-vls) then select the option from the drop down list. Any questions first check your license.dat file or Email support@molsoft.com.
- Click OK IMPORTANT - IF THE JOB IS RUNNING IT WILL TELL YOU bgrnd job AT THE TOP OF THE GUI - SEE BELOW
Docking simulation is running
![]()
Docking simulation has ended message
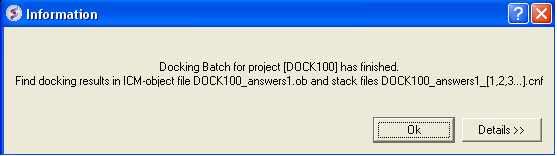
To check the status of your docking simulation
- Windows/Background Jobs
12.3.4 Dock Chemical Table |
To dock directly from a chemical table:
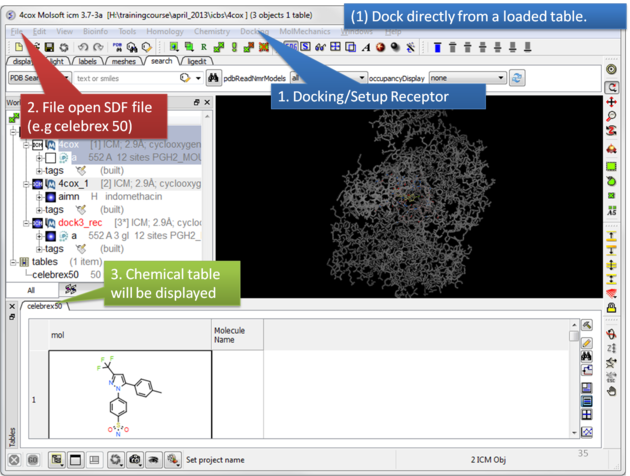 |
| Setup the receptor and read in the chemical table. |
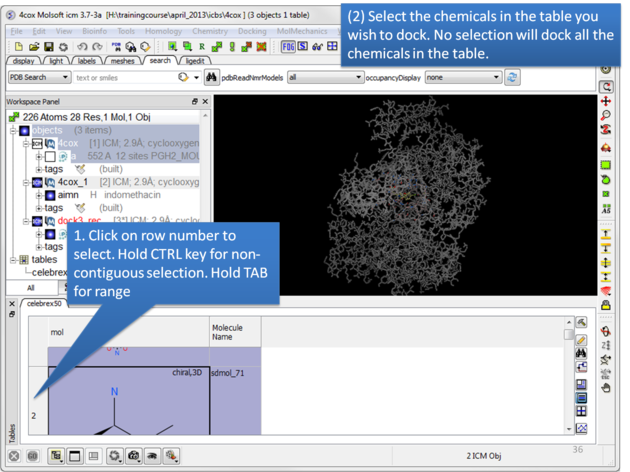 |
| Select the chemicals in the table you wish to dock. If no selection is made ICM will dock the whole table. You can make selections by clicking on the row numbers whilst holding CTRL (non-contiguous selection) or TAB (contiguous selection). |
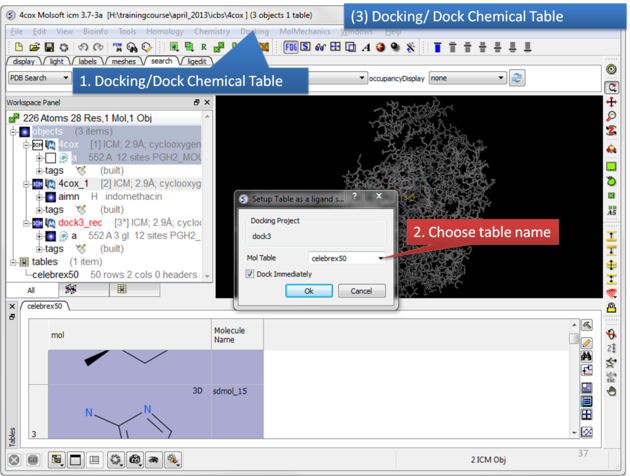 |
|
Click on the Docking menu and select Dock Chemical Table.
To change the default docking options, uncheck Dock Immediately. This will display a panel next to the chemical table where you can adjust settings, such as increasing the number of conformations to save. Then click Dock in the tools panel.
|
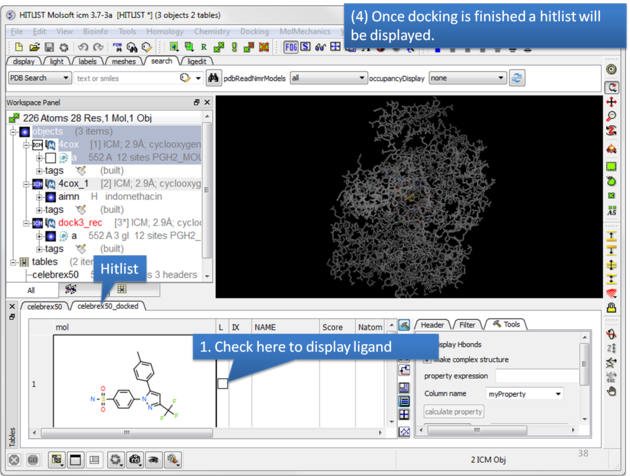 |
| Once the docking has finished a hitlist will be displayed. |
| NOTE: If you want to run the docking again you need to uncheck the boxes in the 'docked' column in your original docking table. To uncheck all - right click on the 'docked' column header and choose 'uncheck'. Then click Dock in the tools panel. |
| Prev Display Preferences | Home Up | Next Display & Analyze Docking Results |