Google Search the Manual:
Keyword Search:
| Prev | ICM User's Guide 9.4 Design Loop | Next |
| Available in the following product(s): Homology |
To design a loop between a gap in your protein structure use the MolMechanics/Design Loop option. This option will search for loops in the PDB which have a similar gap distance and geometry and output energetically favorable solutions detailing which PDB the loop was taken from. If you already have a loop and want to sample it use the loop model option.
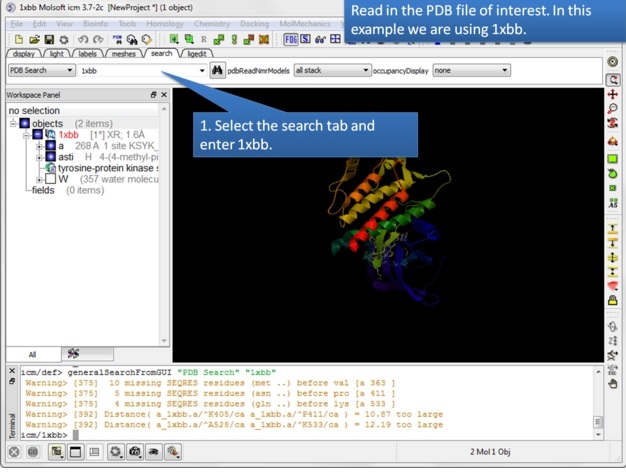 |
| Step 1: Read in the PDB structure. |
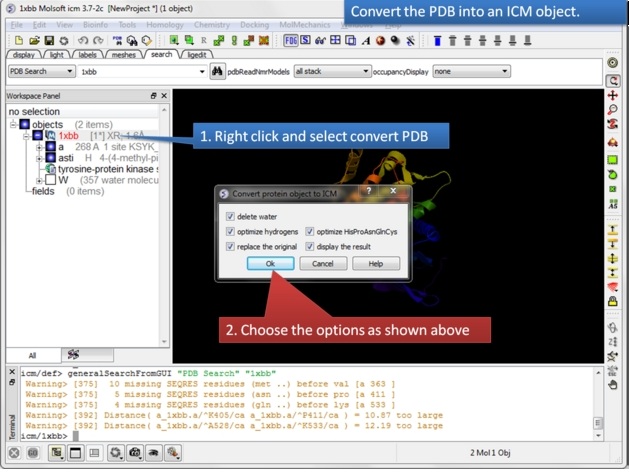 |
| Step 2: Convert the PDB structure to an ICM object. |
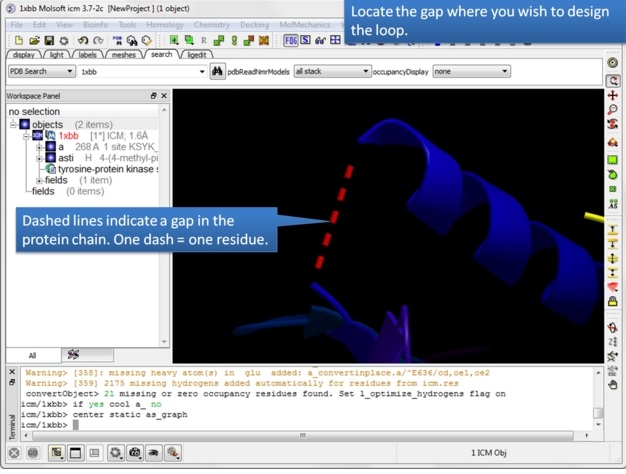 |
| Step 3: Locate the gap where you want to design the loop. Gaps are represented by dashed lines when the structure is displayed in ribbon format. |
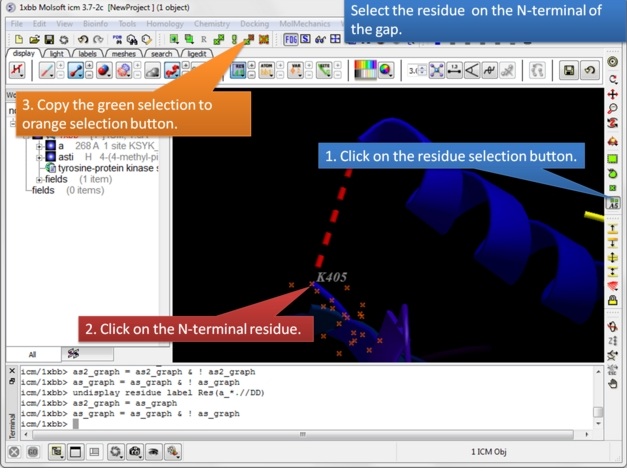 |
| Step 4: Select the N-terminal residue and copy the green selection to orange. |
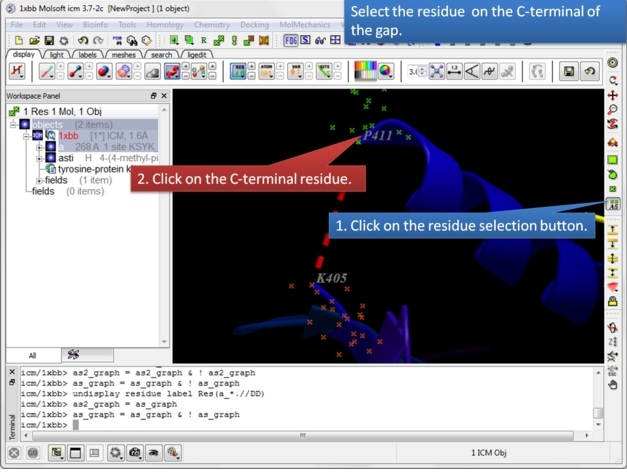 |
| Step 5: Select the C-terminal residue. |
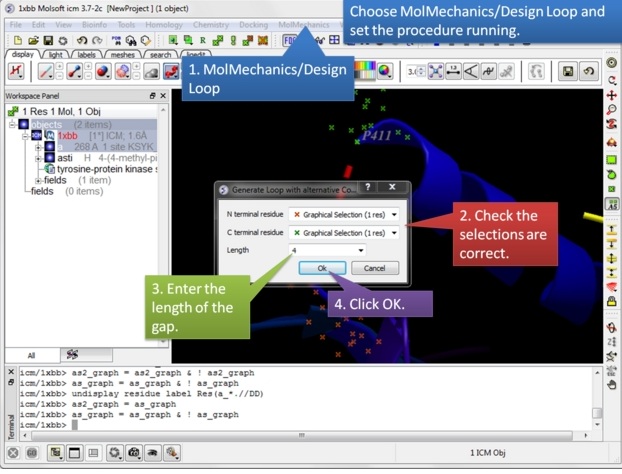 |
| Step 6: Choose MolMechanics/Design Loop menu option. |
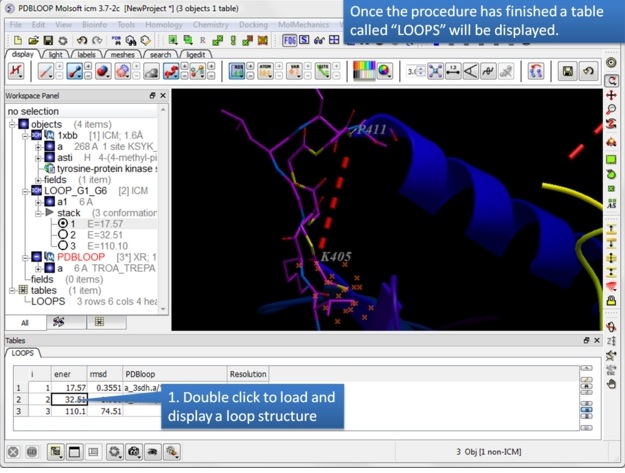 |
Step 7: A table called LOOPS with the loop results will be displayed. Double click on the table to load the loop or use the stack display options in the ICM workspace.
The loops are ranked by energy in the table and the column names refer to:
|
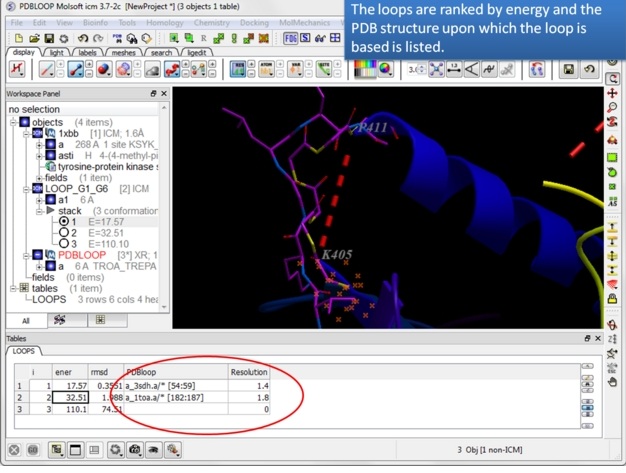 |
| Step 8: Note the PDB upon which the loop design is based is listed in the table. |
| Prev Loop Modeling | Home Up | Next Graft Loop |