iMolView
iMolview is an app for the iPhone/iPad and Android that lets you browse protein, DNA, and drug molecules in 3D. The app has a direct link to the Protein Data Bank (PDB) and DrugBank and has a fast and easy to use interface.
Free Online Tools and Software
Product Feature Comparison
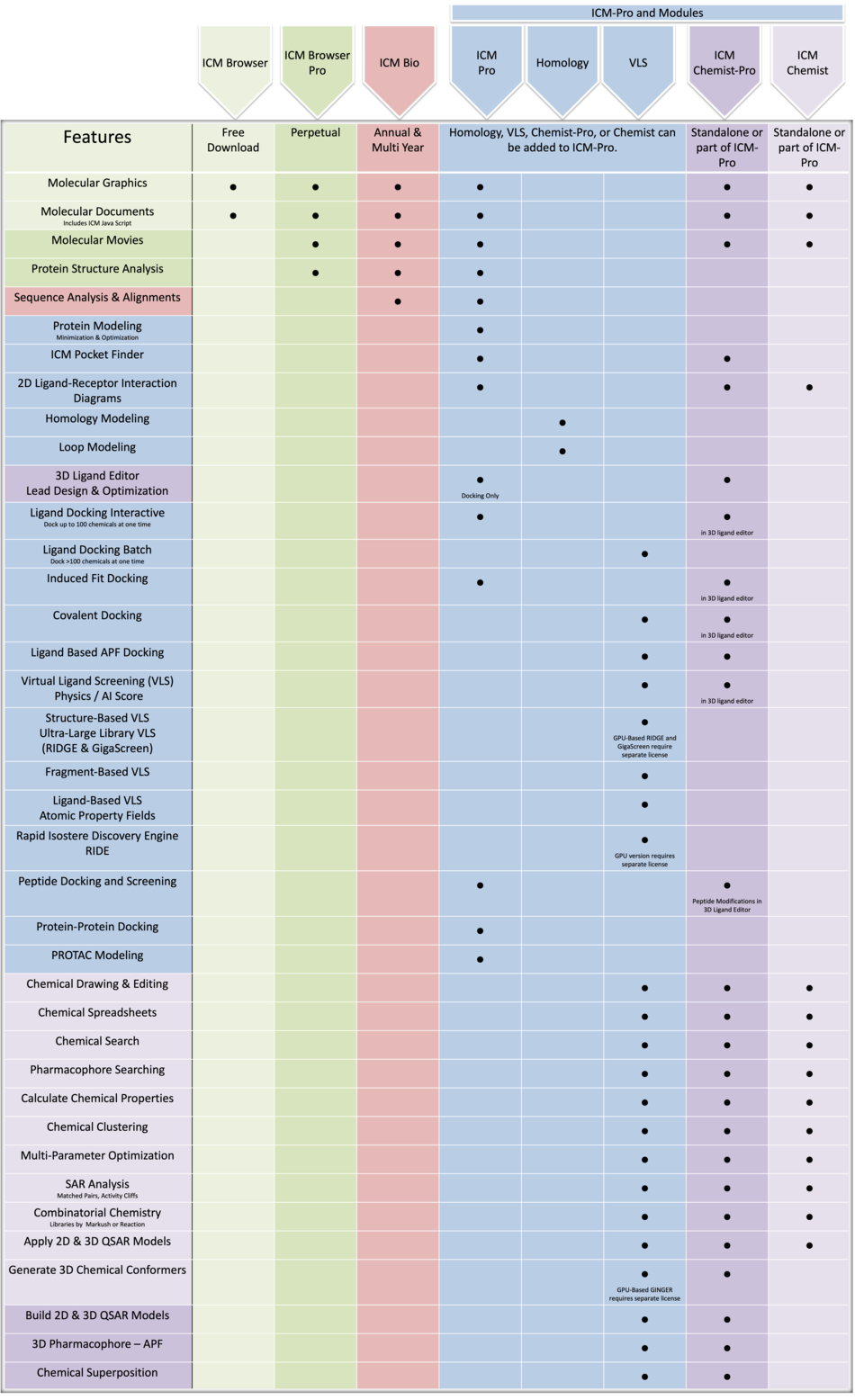
We do our best to keep this list as accurate as possible but please contact us if you have any questions about any particular features and we will be happy to answer any questions you have. Please click here for a more detailed feature comparison.
Licensing
ICM licenses are managed using industry-standard license managers such as FlexLM or LMX.
This allows for flexible control over how the software is accessed and used within an
organization. There are two main types of licenses available:
Floating The license is installed on a central server and managed by the license
manager. Any computer connected to that server can check out the license, provided a seat
is available. This setup is convenient for groups or labs, because it allows multiple
users to access the software from different machines at different times without having
to install a license on each workstation individually. The license manager keeps track
of availability and ensures that the number of concurrent users does not exceed the
purchased seats.
Node Locked The license is bound to a specific computer and managed locally by
the license manager. Only that machine can run the software. This option is typically
used by individual researchers or single-user environments where access from multiple
computers is not required.
Both Floating and Node Locked licenses are offered at the same price. The choice depends
on the intended use: Floating licenses provide greater flexibility in collaborative
environments, while Node Locked licenses are a straightforward solution for single users.
We offer flexible licensing options including free perpetual (ICM-Browser), annual, multiyear, perpetual, site, and teaching licenses.
CPU and GPU Cluster and Cloud Licenses
For high-throughput structure-based or ligand-based virtual screening and molecular modeling,
it is usually more efficient to run the calculations on a dedicated server or through
cloud. The license manager (FlexLM or LMX) handles distribution of
the license across the computing resources, ensuring that jobs can be run in parallel
while respecting the number of CPUs or GPUs licensed.
Unlock Enhanced Modeling Performance with GPU Licensing in ICM
At Molsoft, we understand the importance of optimizing your workflow by harnessing the power of GPUs for specific tools. Regular ICM licenses run on CPUs but our GPU licensing offers a range of specialized license keys tailored to your needs:
Molecular Dynamics: With your regular ICM-Pro license, you can utilize Molecular Dynamics on any available GPU(s) within your computer, unlocking exceptional performance for your simulations. No specific GPU license is required for MD, making it even easier to dive into the world of GPU-accelerated simulations.
RIDE: While RIDE naturally runs on CPU, our exclusive RIDE-GPU license empowers you to accelerate ligand-based screening processes and fully exploit the potential of GPUs. Experience the speed and efficiency gains as you screen millions of chemicals/sec.
RIDGE: RIDGE is designed to fully harness the capabilities of GPUs for structure-based virtual screening. To access its cutting-edge features, acquire a separate RIDGE GPU license key and increase your screening capability more than 100 fold.
GINGER: GINGER (Graph Internal-coordinate Neural-network conformer Generator with Energy Refinement) is Molsoft's new cutting-edge software designed for lightning-fast high quality conformer library generation on GPUs.
Please note at present RIDE, RIDGE and GINGER GPU usage is available to users operating on the Windows or Linux platforms and GPU MD is available on all platforms.
Minimum Recommended Hardware Specifications for CPU Modules
Supported Platforms
ICM runs on the Windows, Mac and Linux operating systems as well as high performance multi-processor clusters and Cloud (AWS, Azure and Google).
- Linux: Any modern Linux distribution will work. At MolSoft we use the following versions (or higher): CentOS release 8, openSUSE 15 and Ubuntu 18
- Windows: All Windows versions are supported.
- Mac: All MacOS versions are supported.
CPU Hardware Specification
- Disk space: At least 2GB of free disk space is required for installation. Additional space may be needed depending on the size of molecular databases, virtual libraries, and user-generated project files.
- Memory (RAM): The software requires a minimum of 2 GB of RAM to run. However, 4 GB is strongly recommended as a default for smoother performance.
- Processor (CPU): A modern multi-core processor is recommended. While the software will run on older CPUs, faster processors with multiple cores will significantly reduce calculation times.
Graphics Card
The Graphic card should have Hardware OpenGL acceleration and 512Mb or more memory. (1024Mb or more is recommended)
We recommend NVIDIA (http://www.nvidia.com) brand.
- GeForce models are good if you do not plan to use hardware stereo.
- Quadro models can be used with hardware stereo
Hardware Requirements for GPU Modules
The following products are optimized to run on GPU using NVIDIA chips on Windows and Linux:
- GINGER - Fast neural network conformer generattion
- RIDGE - GPU enhanced structure-based ligand docking and screening.
- CombiRIDGE - CombiRIDGE rapidly screens combinatorial libraries by growing core fragments with diverse R-groups and ranking them using AI-based scoring.
- GigaScreen - deep learning virtual screening.
- Molecular Dynamics in ICM-Pro.
For all our GPU tools we recommend:
- RTX 3090 or higher (or workstation equivalent) 24Gb RAM or more
- CPU 8 cores or more (16 recommended)
- RAM 64G (128 recommended)
Disk space depends on what you are going to screen the .molt files take up approximately 1 GB per 1 billion chemicals e.g Enamine CombiRIDGE anchor library is 48B chemicals in size and takes 55Gb of disk space.
| Architecture |
Gaming GPUs |
Workstation GPUs |
Data Center GPUs |
| Ampere |
RTX 3090 / 24GB
~$1,000 |
A6000 / 48GB
~$4,000 |
A100 / 80GB
~$8,000 |
| Ada Hopper |
RTX 4090 / 24GB
~$1,600 |
RTX 6000 Ada / 80GB
~$8,000 |
L40CNX / H100
>$30,000 |
Stereo Glasses and Monitors
We recommend the Zalman 3D monitor and Crystal Eye Glasses. Anaglyph stereo is supported in version 3.8 and above therefore only cheap red/cyan glasses are needed (see http://www.molsoft.com/news.html#anaglyph3D ).