
Copyright © 2026, Molsoft LLC
Feb 18 2026
Google Search:
Keyword Search:| Prev | ICM Language Reference O-R | Next |
[ Object | only | parray | pattern | png | Pdb | peptide bond | pharmacophore | profile | prosite | rarray | Psa | REBEL | real | regularization | residue | rgb | ribbon ]
object, 3D molecular objects |
A pdb file or a chemical molecule with 3D coordinates, or a newly build group of molecules that can co-exist in space are called objects in ICM. Please do not confuse this specific use of the word with general shell objects that include other data types. A session may contain any number of molecular objects.Each object (i.e. molecular object) in turn consists of three levels or organization:
- molecules
- residues
- atoms
ICM understands expressions specifying different selections in objects, see object selections. There are two main classes of objects:
- ICM objects
- non-ICM objects
The non-ICM objects (from the second line on) may have subtypes such as
| "ICM" ready for energy calculations. Those objects are either built in ICM or converted to the ICM-type. |
| "X-Ray" determined by X-ray diffraction |
| "NMR" determined by NMR |
| "Model" theoretical model (watch out!) |
| "Electron" determined by electron diffraction |
| "Fiber" determined by fiber diffraction |
| "Fluorescence" determined by fluorescence transfer |
| "Neutron" determined by neutron diffraction |
| "Ca-trace" upon reading a pdb, ICM determines if an object is just a Ca-trace. |
| "Simplified" special object type for protein folding games. |
Each object contains the following information:
- Molecules (with their own internal structure, including residues and atoms)
- name ( changed by rename and returned by the Name ( os ) function )
- remark (set by set comment and returned by the Namex ( os ) function )
- header ( returned by the Header (~~os ) function )
- filename ( returned by the File( os ) function )
- date ( return by the Date function )
- resolution ( returned by the Resolution function )
- type (returned by the Type ( os 2) function )
Functions on molecular objects. os_ means object selection for other abbreviations see the link.(note that some functions that also operate molecules, residues and atoms are not included).
`Cell ( { os | m_map } )
`Charge ( { os | ms | rs | as } [ formal | mmff ] )
`Date ( os )
`Field( { rs | ms | os } [ i_fieldNumber ] )
`Field( { as | rs | ms | os } [ s_fieldName ] )
`Field ( os i_fieldNumber ) # see also set field os_ i_number
`File ( os )
`Label ( os_objects )
`Mass( as | rs | ms | os )
`MaxHKL( { map | os | [ R_6CellParameters ] }, r_minResolution ) → I3
`Mol ( { os | rs | as } )
`Name ( s_hint os unique )
`Name ( os )
`Name ( s_root os_1 unique )
`Namex ( os )
`Next ( { as | rs | ms | os } )
`Nof( { os | ms | rs | as | vs } )
`Nof( { os | ms | rs } [ atom ] )
`Parray ( object [os] )
`Res ( { os | ms | rs | as } [ append ] )
`Resolution ( os_object ) → R_array_of_resolutions
`Select ( as_sourceSelection os_targetObject )
`Select ( os_sourceObject S_residueSelStrings )
`Select ( os_sourceObject I_atomNumbers )
`String( { os | ms | rs | as } [ i_number ] )
`Symgroup ( { s_groupName | os_object | m_map } )
`Symgroup ( { i_groupNumber | os_object | m_map } string )
`Transform( s_group|iGroup|os_1|map ) → R_12N_all_fract_transformations
`Transform( s_group|iGroup|os_1|map iTrans ) → R_12_fract_transformation_i
`Type ( os_object , 2 )
`dsCellBox os_
Commands acting on objects
append stack os_ligandObject
clear graphic [ os_ ]
convert [ exact ] [ charge ] [graphic] [ tether ] [ os_non-ICM-object ] [ s_newObjectName ]
copy [ strip ] [ tether ] os [ s_newObjectName ] [ delete ]
delete { object | os }
find pdb rs_fragment os_objectWhereToSearch s_3D_align_mask [ s_sequencePattern [ s_SecStructPattern ] ] [ r_RMSD_tolerance]
find pattern [ number ] [ mute ] s_sequencePattern [ i_mnHits] [ { os_objectWhereToSearch | seq_Name | s_seqNamePattern } ... ]
find segment ms_source [ os_objectWhereToSearch_2] [ r_maxRMSD ] [ i_minNofAlignedResidues [ i_accuracyParam ] ]
move ms_MoleculesToMove os_destination
move os_ObjectToMove os_destination
rename { os [ full ] | ms | rs | as } s_newName
rotate [ os | ms | g_grob ] M_rotation
set comment [ append ] os_Object s_comment # returned by the `Label() function
set object [ os_newObjName ]
set symmetry os_object R_6cell s_symgroup | i_symgroup [ i_NofChains ] # set symmetry os s_crysym_record
set type os s_type
set [ atom ] type mmff [ os]
show { os | ms | rs | as | vs }
sort object R|I # reorder objects by arrays, e.g. --{sort object Resolution(a_*.)}
sort object os_ i_pos # move selected OBJECTS to specified position
sort os [ field = i_Field ] # sort MOLECULES in selected objs by field
strip os_object [ virtual ]
superimpose os_static I_atomNumbers1 os_movable I_atomNumbers2
translate { os | ms | g_grob } [ add ] [ symmetry ] R_3translationVector
Important macros working on molecular objects.
convertObject ms l_delete_water l_optimize_hydrogens l_replace_the_original l_display
convert2Dto3D os l_build_hydrogens (yes) l_fixOmegas (yes) l_display (no) l_overwrite (yes)
icmPmfProfile os ( a_ ) l_accessibilityCorrection (yes) l_display ( no )
makeSimpleModel seq ali os
makeSimpleDockObj [ os_object ] [ s_newObjName ]
only |
frequent option in commands which means disregard or delete the previous status. Without only commands usually add or append to the current settings.
Examples:
display only g_icos # undisplay everything which is in the
# graphics window (if any)
# and display icosahedron
parray |
[ Object parray | Sequence parray | Image parray ]
pointer array, abbreviated as P. An array of pointers to objects. Currently there are several types of parrays, that consist of the following data types:- chemical
- image
- sequence
- distances (e.g. interatomic distances, hbonds, angles, torsions)
- 3d labels
- object
- slide
creating mol-arrays To create a molarray use the read table mol command and first column of this table will be such an array. Example:
read table mol "Maybridge.sdf"You can also use the Chemical function.
Molecular objects in arrays and tables |
ICM molecular objects (both converted and unconverted) can be grouped into arrays and shown in tables.
See the following functions:
Array of sequences |
ICM sequences may be grouped into arrays and placed into tables. Some functions supporting sequence arrays:
- Sequence array
- Parray( object ... )
- Tostring
- Length
- Name sequence
- set name sequence
- Sstructure
- set sstructure sequence
- Namex sequence
Array of images |
ICM provides multiple ways of generating images and performing advanced operations with them: See
- Image
- Info(image)
- make image
- set background image
- set texture
- Sum image
- Color image
- Name image
- Namex image
- image
pattern |
a sequence consensus pattern like this, "[AG]?[!P]W", or this "C?G?\{2,3\}C". A pattern can be extracted from an alignment and searched against a sequence database. Allowed elements:
* # anything until the next element, e.g. Find( "*[!A]?SI*" sequence )
? # any symbol
[XY..] # any one of the listed one-letter codes, e.g. [DE] for asp or glu.
! # negation, e.g. [!PG] anything except P or G
\{l\}, or \{l1,l2\} # anything of length l, or from length l1 to l2, e.g. \{0,5\}
#
# Example:
#
Find( "*A[LV]\{0,5\}*[!DE]?\{8\}C*" sequence )
See also:
- find pattern - find a pattern in a single sequence,
- find database pattern - efficient parallel pattern search in a BLAST-formatted sequence databank.
- Pattern( s_consensus) - create a regular pattern expression from a consensus,
- Pattern( alignment) - create a regular pattern expression from an alignment,
- regular expressions and pattern matching
- prosite - a collection of sequence patterns
- Find( s_pattern sequence ) to find pattern in all loaded sequences
png |
[ pattern-matching ]
graphics image format. Stands for Portable Network Graphics and was designed to replace the GIF format and, to some extent, the much more complex TIFF format. While GIF allows for only 256 palette colors, PNG can handle a variety of color schemes like TIF (1,3,8, 24, etc. bit colors). Furthermore, PNG is free, while GIF is subject to licensing fees. PNG also supports alpha-channel. Since 1998 most browsers correctly display PNG images.To write a png with transparent background use write png transparent
Pattern matching and regular expressions. Use the following metacharacters to construct regular expressions (try guess what string is used in the examples!)
- * matches any string including an empty string (e.g. "*see*" )
- ? matches any single character (e.g. "???ee M")
- [string] matches any one of the enclosed characters. Two characters separated by dash represent a range of characters. Examples: [A-Z], [a-Z], [a-z], [0-9] (e.g. "[A-Z] see [A-Z]"
- [ !string] negation. matches any but the enclosed characters (e.g. "I see [!K]")
- single-character multiplication: character\{m,n\} (e.g. "I?\{3,6\}M" - repeat any character, ?, from 3 to 6 times)
pdb or Protein Data Bank |
a repository of macromolecular structures solved by crystallography or NMR (occasional theoretical models are frowned upon). It used to be at the Brookhaven National Laboratory, Now it is shared between UCSD and Rutgers University. The old citations: Bernstein et al., 1977; Abola et al., 1987). The new citations can be found at http://www.rcsb.org/pdb/ . On November 20th, 2001 it contained 16596 entries. On August 21st, 2007 if contained 45368 Structures.
An example ATOM record:
ATOM 52 N HIS D 18 53.555 24.250 49.573 1.00 32.59
peptide bond |
a covalent bond between C=O and N-H groups, which is imposed in ICM-objects as an extra set of distance restraints. These groups may belong to the terminal groups as to the amino acid side chains. Important: commands make peptide bond and delete peptide bond are valid for ICM-type molecular objects only (and have no effect on, say, PDB structures). Both commands change the covalent structure of the modeled molecular object and expel/add hydrogens. Distance restraints imposed to form such a bond are defined in icm.cnt file.
pharmacophore |
Pharmacophores in ICM are special objects of the non-ICM type. A pharmacophore can combine special ph4 centers and disconnected chemical fragments.
The ph4 centers have the following atom types and names:
- code:
- 315 Qa HBA
- 316 Qd HBD
- 317 Qm aromatic
- 318 Qh hydrophobic
- 319 Qp positive
- 320 Qn negative
- 321 Qx ExVolume
- 322 Qv a point defining the direction vector from Qa,Qd and Qm, or a pseudo atom
Creating a pharmacophoreA pharmacophore can be created from an object with the makePharma macro,
macro makePharma ms s_name ("pharm") l_points (yes) l_display
The l_points logical defines if the pharmacophoric centers or fragments are created from the source molecule.
The new object gets type pharmacophore (see the set type os "pharmacophore" command ).
Note the following issues:
- for polar atoms with hydrogens and lone pairs, both Qa and Qv centers are created at the s
- the directions of Qa-Qv or Qd-Qv vectors need adjustment
- the direction atoms (Qv) can be selected and removed. In this case the direction will not be matched.
- two or more ph4 objects can be merged into a single pharmacophore object. For example one can merge some ph4-centers with some fragments.
- the properties of points can be edited interactively or with the set as_ commands.
- set bfactor as1_ r_radius # to change the tolerance radius
set bfactor a_pharma.p5/5/Qv10 0.8
- resetting the radius (stored in the bfactor field) for the direction point Qv will change the width of the direction tolerance.
- you can also change point type and name, e.g.
set type a_pharma.//Qa3 316 rename a_pharma.//Qa3 "Qd4"
- the coordinates can be reset with the set atom command.
The following commands and functions work with pharmacophores:
- find pharmacophore : find pharmacophore as_pharmQuery chemarray3D
- superimpose : superimpose os_ph4obj ms_ligand pharmacophore
- Chemical : Chemical( ms|os pharmacophore ) # creates pharmacophore, this function is used in makePharma
- Rmsd : Rmsd ( as_pharmTemplate as_select2 pharmacophore )
- makePharma macro
- makePharmaAtomsmacro # generates separate pharma centers from selections
Saving ph4 objects.
The objects can be saved with the regular write object command or write binary commands.
profile |
a table of residue preferences for each residue type at each position on a protein fold or a sequence. The preferences may be derived from a multiple sequence alignment of from a 3D structure. Profile also contains gap opening and gap extension values for each sequence position. Profile provides a good way of representing a consensus sequence pattern of a protein family. One can search a new sequence against a library of profiles, or search a profile against a data base of protein sequences (see Abagyan, Frishman, and Argos, 1994). One can add two profiles ( prf1 + prf2 ), multiply them (prf1 * prf2), concatenate two profiles (prf1//prf2), and extract a part of a profile ( prf[15:67] ). Profile can be read from a .prf file and calculated from an alignment with the Profile() function. To make a table out of a profile use this example:
make sequence 10; align sequence; prf=Profile(aln);
t = Table(Matrix(prf)) # columns names 'A','B','C',...
add column t Count(Nof(t)) Split(String(Sequence(prf)),"") name={"ipos","seq"} # adds number and cons seq
See also:
Sequence()
Consensus()
Matrix()
Align().
prosite |
a dictionary of protein sites and patterns distributed by the Swiss Institute of Bioinformatics, http://ca.expasy.org/prosite/ , Copyright by by the Swiss Institute of Bioinformatics (SIB). it is also one of possible formats for string patterns. ICM converts prosite patterns to standard string patterns containing regular expressions, like "C?\{4,5\}CCS??G?CG????[FYW]C".
Included in ICM is only the old release 13 (1995). Until the 15th release the files were fully open for redistribution and said this.
CC ************************************************************************* CC CC This file may be copied and redistributed freely, without advance CC permission. You are allowed to reformat it for use with a software CC package, but you should not modify its content without permission CC from the author). CC CC *************************************************************************.
If you want to use a newer version of prosite and your organization is non-profit or has a license from SIB, you can download prosite.dat from, e.g.:
unix wget ftp://us.expasy.org/databases/prosite/release_with_updates/prosite.dat read prosite "./prosite.dat" # read the newer version in readUniprot "IL2_HUMAN" find prosite IL2_HUMAN
See also:
- read prosite,
- s_prositeDat,
- find prosite - find all prosite patterns in a single sequence,
- find profile - find all prosite profiles in a single sequence
rarray |
ICM class for arrays of real numbers: { -1.6 , 2.150 3., -160.} An array can be formed dynamically from simple reals with the double slash (concatenate) operator, e.g. 1.3//-1.//dielConst//14.
Real arrays also support special values (see rarray constant )
- ND (not defined)
- >r_value
- <r_value
- INF : infinity
- -INF : infinity
Example:
add column t {1.}//ND//Real(">2.")//Real("<3.")//Real("INF")//Real("-INF")//2.3
add column t
See also: Rarray , Real , Toreal , read rarray , rarray constant , GUI.displayNDValueStyle
Predicting Polar Surface Area |
Polar surface area or PSA is a parameter important for predicting transport properties of drugs. It correlates with intestinal absorption and blood brain barrier penetration. (e.g. see Polar Surface Area by Peter Ertl in Molecular drug properties: measurement and prediction, Raimund Mannhold, Ed, Wiley-VCH, v 37, 2008 )
Single conformation 3D PSAPSA is calculated as the exposed van der Waals surface of oxygens and nitrogens as well as the hydrogen atoms attached to them. For a converted and current ICM object PSA can be calculated with the following command:
show surface area a_//n*,o* | ( a_//h* & Next( a_//n*,o* bond )) a_ mute vwExpand = 0. PSA = r_out # the previous command returns the value in r_out
The conversion is needed only to make sure that all hydrogens are in place. If they are, the conversion is not really needed and the same command can be applied.
Multiple conformation 3D PSAThis measure obviously depends on the conformation of the molecule. An average PSA can be calculated with the following script from a stack on conformers generated by a montecarlo command, e.g.:
for iconf=1,Nof(conf)
load conf iconf
show surface area a_//n*,o* | ( a_//h* & Next( a_//n*,o* bond )) a_ mute vwExpand = 0.
avPSA += r_out
endfor
avPSA /= Nof(conf)
This script can also be easily modified to include Boltzmann weights of individual conformer contributions.
A similar look can be written to process the results of the _confGen script.
Fast and accurate estimate of PSA from chemical topology only ( TPSA or MolPSA )
Ertl et al. (see the reference above) proposed a way to estimate the PSA value directly from a two dimensional topology of a molecule without a need to convert it to 3D and sample its geometrical states. The ICM MolPSA function uses a similar method to estimate PSA, ( e.g. Predict( Chemical("CCO"),"MolPSA" ) ). The following method was used to derive a model (see the learn command ):
- Six thousand drug compounds were converted to 3D, optimized and their real single conformation PSA values were evaluated
- The chemical topologies were converted into Molsoft fingerprints and trained against the computed PSA values
- The resulting model had a cross validated squared correlation coefficient of predicted vs training values of Q2=0.99 and the root mean square error of Rmse=1.56.
read table mol s_icmhome+"/moledit/Dictionary.sdf" name="t" add column t Predict(t.mol "MolPSA" ) name="MolPSA" show Mean(t.MolPSA)
REBEL |
| a method to solve the Poisson equation for a molecule. REBEL is a new powerful implementation of the boundary element method with analytical molecular surface as dielectric boundary. This method is fast (takes seconds for a protein) and accurate. REBEL stands for Rapid Exact-Boundary ELectrostatics. The energy calculated by this method consists of the Coulomb energy and the solvation energy which is returned in the r_out system variable. |
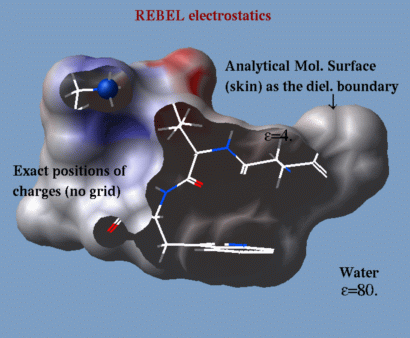
|
- electroMethod = "boundary element";
- .`TOOLS.rebelPatchSize (1., but can be smaller, say 0.3 or 0.5, it is a tradeoff between speed and accuracy)
- dielConst (the default is usually OK);
- dielConstExtern (the default is usually OK);
- set charge as_ r_Charge (modify charges if you like);
- make boundary (if you want to make several evaluations of energy or Potential( ) with the same boundary. The "boundary" parameters depend only on conformation and do NOT depend on charges. You can redefine charges afterwards and get a corrects energy evaluation);
- delete boundary (if you do not need it);
- show energy (make sure the "el" term is on);
- Potential ( as_targets as_charges) (if the boundary exists, returns potentials from charges at the target atoms);
- color grob potential [heavy] (create graphics object, say, with make grob skin actually it can be any grob and color it by the REBEL potential); option heavy results in better coloring.
- Matrix(boundary) : returns the matrix solved to find the boundary element charges.
- dsRebel : a macro for calculating the potential-colored electrostatic surface.
The polarization charges, or induced surface charges, can be returned by the Rarray( as_ ) function after the equation is solved, e.g.:
electroMethod = 4 show energy "el" Rarray( a_//* ) # returns polarization charges
Electrostatic troubleshooting
Problem: the electrostatic surface looks different before and after conversion to ICM .
Explanation: The unconverted PDB object has (i) an incomplete set of atoms ( lacking hydrogens for the X-ray entries, occasional missing side chains or loops, etc.); (ii) has formal charge ambiguities for his (hip), asp (ash) etc. , see below (iii) has no charges, i.e. the formal and partial charges are not assigned;
The dsRebel procedure (i) builds a full atomic model with hydrogens, (ii) resolves at least some residue charge ambiguities, and (iii) assigns to the atoms of the unconverted objects a simplified set of formal charges. See convertObject for further details.
The difference between the formal charges of the unconverted and the full distributed partial charges after conversion creates a small difference in electrostatic pattern.
If the conversion procedure charges a residue (e.g. a cysteine next to a metal ion), then the electrostatic patterns will differ much more in the vicinity of that residue.
Problem: the electrostatic surface looks different for different PDBs of the same protein
Explanation 1: Some residues can be charged or uncharged. In a specific PDB can be marked by a different 3-letter residue name or not. For example:
| Amino Acid | Frequent | Rare |
|---|---|---|
| aspartate | asp (-) | ash (0) |
| cysteine | cys (0) | cym (-) |
| glutamate | glu (-) | glh (0) |
| histidine | his (0) | hip (+) |
| lysine | lys (+) | lyn (0) |
| tyrosine | tyr (0) | tym (-) |
Problem: the numerical singularities are reported Sometimes numerical instabilities inherent to the REBEL calculation lead to singularities and strange values of electrostatic potential. This can be detected by looking at the polarization charge values. The ePotProtected macro in the _rebel file contains the code that checks for this type of instabilities.
real |
number may exist in the ICM-shell as a named variable or a constant (e.g. 12.3, 2.0, -4.501 ). There are a number of predefined real variables. Reals can be concatenated into a real array like this: 12.3//2.0//-4.5
Reals may be mentioned in
arithmetic expressions,
commands and
functions.
Examples:
a = -1.2 b = Abs(Sin( 2.3 * a - 3.0 / a))
regularization |
procedure for fitting a protein model with the ideal covalent geometry of residues (as represented in the icm.res residue library) to the atom positions of a target PDB structure (usually provided by X-ray crystallography or NMR). Regularization is required because the experimentally determined PDB-structures often lack hydrogen atoms and positional errors may result in the unrealistic van der Waals energy even if these structures were energetically refined (since the refinement of the crystallographic structures typically ignores hydrogen atoms and employs different force fields). The following steps are required to create the regularized and energy refined ICM-model of an experimental structure:
- an extended all-atom model of a particular protein is generated with regular geometry characteristics (see the build command and the IcmSequence function);
- the non-hydrogen atoms in the model are assigned to the equivalent atoms in the model (see set tether);
- the regularized structure is built starting from the N-terminus by adding atoms one-by-one (see minimize tether);
- methyl groups are rotated to reduce van-der-Waals clashes;
- combined geometry and energy function is optimized;
- polar hydrogen positions are adjusted;
- optionally the model may be additionally minimized, now without tethers to observe a "stability" of the model in the local energy minimum.
residue |
a chemical building block or complete chemical compound, usually an amino-acid residue. The ICM hierarchy: atom -> residue -> molecule -> object. Individual small molecules may contain only one residue. Residues are described in the icm.res file. You may create your own residues with the write library command. Residues can be selected with the ICM-selection expression (e.g. a_/ala, a_/15, a_/15:20, a_/"RDGE" etc.), labeled with the display residue label rs_ command, by double clicking with the right mouse button, via a pop-up menu, or from * Added tothe registry{ Table( model chem [inverse] ) } function. the GUI menu.
rgb |
red-green-blue. It is of interest, that the combination of these three can produce any other color. In addition, this is the name of the SGI image format used in the ICM commands write image and display trajectory . ICM also generates the fourth channel on top of the RGB information. This fourth number is called alpha-channel and generates the opacity index for each pixel of the image. This information is interpreted by a number of applications, i.e. the IRIX showcase and dmconvert (the SGI moviemaker). See also tif, targa, postscript.
ribbon |
|
a graphical representation of a polypeptide chain backbone by a smooth
solid ribbon. DNA and RNA can be also displayed in a ribbon style.
There are three types of elements of the ribbon display depending on the secondary structure assigned to a given residue. |

|
Residues marked as alpha-helices ('H') will be shown by a flat ribbon, those marked as beta-sheets ('E') will be flat ribbon with an arrow-head, and the rest will be shown by a cylindrical "worm". The ICM-shell parameter GRAPHICS.wormRadius defines its radius. Default ribbon colors are defined in the icm.clr file. Note that minor secondary structure elements like 3/10 helix ('G'), Pi-helix ('I') are colored by the corresponding colors ( the threetenRibbn and piRibbon parameters in the icm.clr file), 'Y' type is colored by the alphaRibbon color, and 'L','P' and 'B' (isolated beta-residue) residues are colored by the "betaRibbon" color. DNA and RNA ribbons are colored according to the base type: A-red, C-cyan, G-blue, T or U - gold. Preference ribbonStyle allows one to display a simplified segment representation of the secondary structure elements instead of (or together with) the ribbon.
If the polypeptide chain has a break (missing residues) the ribbon
shows bullets or nothing depending on the GRAPHICS.chainBreakStyle .
The DNA/RNA ribbons consists of two parts the backbone ribbon
and the bases shown with the sticks and balls. To selectively
display and undisplay the bases, you can do the following:
Example:
read pdb "1dnk" # contains 2 dna mol. display ribbon a_1.2,3 # both bases and backbone undisplay ribbon base a_1.2 # bases disappear display ribbon base only a_1.2 # only bases display ribbon a_1.2,3 yellow # both bases and backbone color ribbon a_1.3 magenta # the second chain backbone color ribbon a_1.2,3 bases # default by base type cool a_ # cool is a rich macro. View the whole thingSee also:
- display ribbon command (also undisplay and color )
- GRAPHICS.ribbonRatio parameter
- GRAPHICS.ribbonWidth parameter
- GRAPHICS.ribbonCylinderRadius parameter
- GRAPHICS.dnaRibbonRatio parameter
- GRAPHICS.dnaRibbonWidth parameter
- GRAPHICS.ribbonGapDistance parameter
- GRAPHICS.dnaRibbonWorm logical
- GRAPHICS.ribbonWorm logical
- l_breakRibbon logical
- GRAPHICS.chainBreakLabelDisplay preference
- GRAPHICS.chainBreakStyle preference
- GRAPHICS.dnaRibbonStyle preference
- ribbonColorStyle preference
- ribbonStyle preference
- residue field "_WORMRADIUS_" for variable thickness
| Prev mute | Home Up | Next S |