| Prev | ICM User's Guide 9.4 Graft Loop | Next |
| Available in the following product(s): Homology |
To transfer a loop from one object to another:
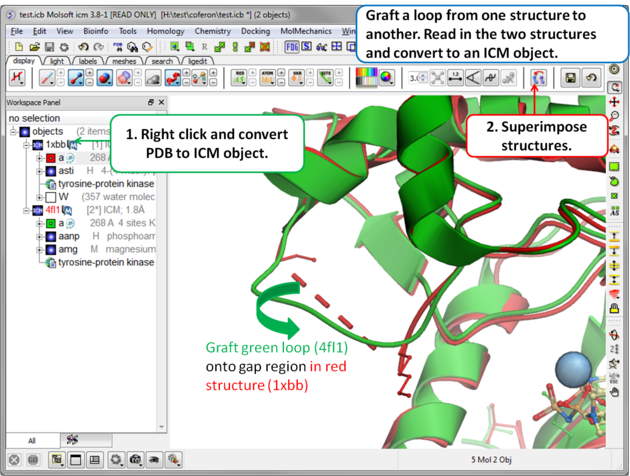 |
Step 1: Read in the structures and convert to ICM objects.
|
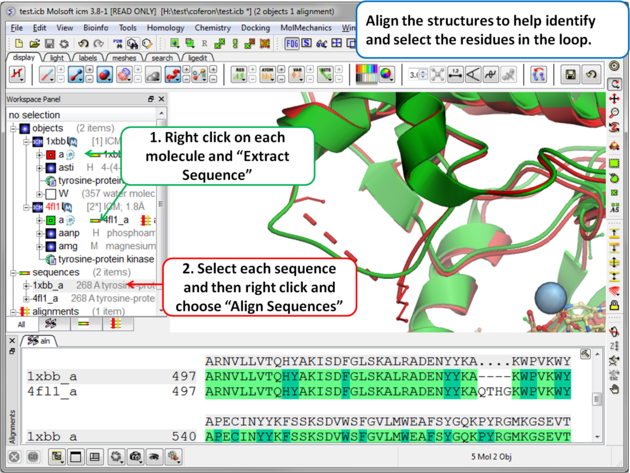 |
Step 2 (Optional): To help identify and select the gap region you can align the PDB sequences.
|
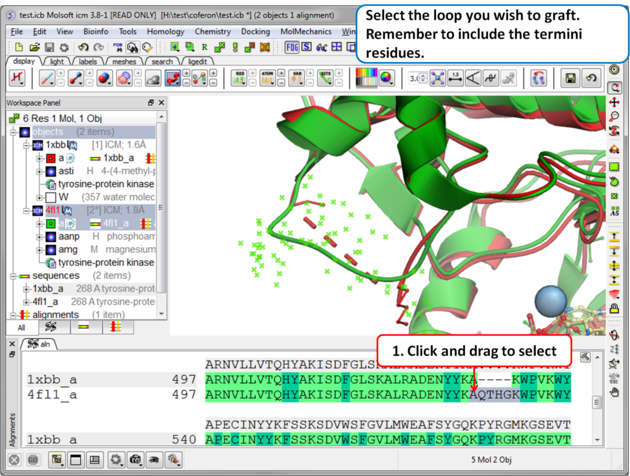 |
Step 3: Select the source loop.
|
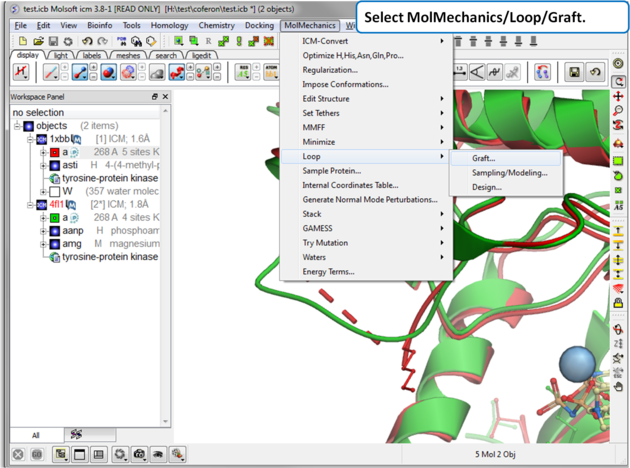 |
| Step 4: Select MolMechanics/Loop/Graft |
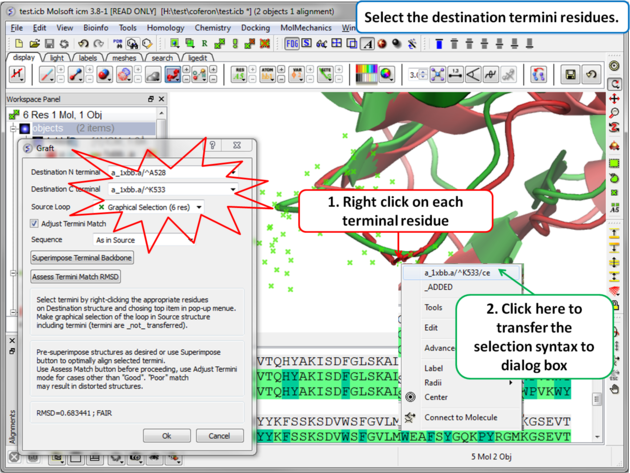 |
Step 5: Select the termini residues of the structure with the gap (destination).
|
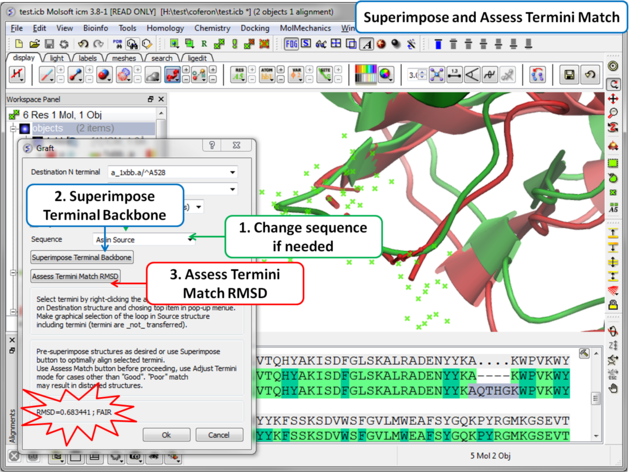 |
| Step 6: Superimpose termini backbone and assess the quality of the match. |
- You can change the sequence of the residues in the grafted gap (click on the drop down button in the Sequence dialog box).
- Click on the superimpose terminal backbone button.
- Use "Assess Match" button before proceeding, the RMSD and report will be displayed in the terminal box. Use adjust termini mode for cases other than Good.
- Click OK.
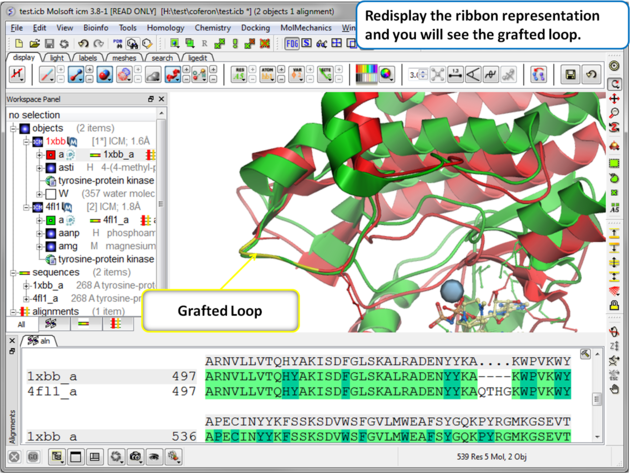 |
| Step 7: Display the grafted structure. Sometimes you may need to refresh the ribbon display to view the new loop. |
| Prev Design Loop | Home Up | Next Loop Preferred Residues |