
Copyright © 2026, Molsoft LLC
Feb 18 2026
Google Search:
Keyword Search:| Prev | ICM Language Reference Other shell variables | Next |
[ defCell | accFunction | gapFunction | I_out | M_out | R_out | R_2out | S_out | S_proteinTags | swissFields | readMolNames | Selection variable | as_out | as2_out | vs_out ]
defCell |
the real array of the default cell parameters. This definition is used in the Resolution and MaxHKL functions if cell parameters are not provided as arguments.
Default: {1. 1. 1. 90. 90. 90.}
accFunction |
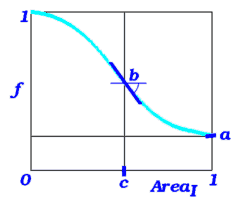
|
the real array of the solvent accessibility penalty parameters (as described in Batalov and Abagyan, 1999). It contains the values of a, b, c and E damping parameters for amino acid substitution scores. Generally, if a residue is completely buried ( Area=0), its substitution scores will be used without changes. If it is completely exposed, its substitution scores will be multiplied by the minimal possible value of a. Between these cases the substitution scores are modulated by a smooth ("arctangent") function with a saddle point at Area=c, where the slope will be -b. The fourth parameter is reserved for development. |
This definition is effectively implemented in the Align( seq_1 seq_2 area ) }, Score functions and find database command.
Default: {0.33 2.35 0.211, -15.0}.
See also: alignMethod .
gapFunction |
the real array of the gap penalty parameters, which represent a piecewise-linear concave function (as described in Batalov and Abagyan, 1999).
ATTENTION: at the present time this gapFunction is only active when alignMethod =2.
The first two values replace gapOpen and gapExtension traditional values. If present, the third element of the array represents the length of the gap, starting at which further gapExtensions become equal to the fourth element of the array. Likewise, if more elements are present, they represent pairs of the threshold lengths of the gap and the new gapExtensions values. For example,
gapFunction = {2.4 0.15 10. 0.05 20. 0.}
means that
- gap penalty=2.25+0.15*L for L={ 0..10} (and for L=1 it is 2.4= gapOpen ),
- gap penalty=3.25+0.05*L for L={11..20} and
- gap penalty=4.25 for L>20
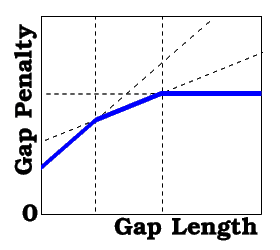
|
The calculations are fastest for the traditional two-element gapFunction. The three- or four-element gapFunction invokes the optimized routines and is 50-70% slower. The general kind gapFunction costs approximately 70-90% additional time for every pair of gapFunction values. If the last gapExtension is zero, it may be omitted. This definition is effectively implemented in the Align , Score functions and find database search command. |
Default: {2.4 0.15}.
Recommended (put it in your _startup file): gapFunction = {2.4 0.15 10.}
This set will produce fast and structure-like alignments.
See also alignMethod, and accFunction (the accessibility attenuation parameters).
I_out |
an integer array in which the output of some commands is stored.
M_out |
matrix in which the output of some commands is stored.
R_out |
real array in which the output of some commands is stored.
Functions returning in R_out:
- Axis # middle point of the axis
- Disgeo # returns error sum of negative scaled eigen values in R[0], and first three 3 scaled eigen values
- LinearFit # residuals
- Xyz(
| .. ) returns inverse transformation | - learn returns model accuracy and stds.
R_2out |
auxiliary rarray. Used in addition to R_out.
S_out |
string array in which the output of some commands is stored.
S_proteinTags |
sarray of sequence patterns that can be used for detection or deleting those tags from a sequence using Trim ( seq S_tags ) function.
swissFields |
string array of SWISS-PROT fields to be read by default in read sequence swiss
If the field name starts from a minus ('-'), this field will be ignored in the feature table list. Also note that variable i_2out (after read sequence) contains the beginning of mature sequence.
Example:
swissFields={"-HELIX ","-COIL ","-STRAND","-TURN "}
# to suppress the FT records with the secondary structure info
readMolNames |
string array in which the SDF-file comment fields containing database compound identifier and description are preset. There is a standard place where database compound identifier should be stored in SDF (MOL)-files. This is the first line of the entry. However most of the database providers got used to leaving this line empty. Instead they put identifier and description in the end of the file in the following fashion:
... M END > <CAT_NO> R150002 > <NAME> (5-OXO-HEXAHYDRO-PYRANO[3,2-B]PYRROL-1-YL)-ACETIC ACID METHYL ESTER $$$$In this particular case before using such database set
readMolNames = {"<CAT_NO>" "<NAME>"} # useful for Sigma-Aldrich files
Another example:
readMolNames = {"<CODE>" "<IUPAC_NAME>"} # useful for ACD database
Named Atom/Residue/Molecule/Object/Variable Selections |
Selections of atoms, residues, molecules, objects or internal variables (torsions, planar angles, bond lengths) can be stored in variables.
Examples:
cc = a_//ca # created named selection variable cc show cc & a_/3:15 # use it in the expressionIn this case the named selection cc is a true ICM-shell variable, not just an alias for the Ca selection. Please do not confuse it with another useful mechanism which allows you to use a string in a selection. This mechanism is used in scripts and macros.
Example:
cc = "a_//ca" # in this case cc is a string, not a selection show $cc & a_/3:15 # $cc is replaced by a_//ca before parsing
How to store and exchange selections in strings:Examples of using the String ( os|ms|rs [name|number] ) function to return a residue selection:
l_showResCodeInSelection = yes # the default res_str = String( Res(Sphere( a_H [1] a_A//!h*,ca,c,n,o 3.5 ))) # same as with option String(.. name) show res_str a_2c0cb.b/^T159,^S205,^K209,^Y224,^V248,^Y275,^L305,^M356,^N361 # or l_showResCodeInSelection = no res_str = String( Res(Sphere( a_H [1] a_A//!h*,ca,c,n,o 3.5 ))) # same as with option String(.. number) show res_str a_2c0cb.b/159,205,209,224,248,275,305,356,361However, be careful with using it in an arbitrary case at the atomic level since it may lead to a string that is too long.
as_out |
an atom/residue/molecule/object selection variable where some commands or functions store their output: If atoms a_1./3/ca,c,n relate to atoms a_2./45/ca,c,n, then the first set will end up in as_out and the second in as2_out.
as2_out |
the second set of atoms ( selection ) returned by the following commands and functions: See also: as_out.
vs_out |
The variable selection where some commands or functions store their output:
- read variable saves a selection of loaded variables;
| Prev WEBAUTOLINK | Home Up | Next Chemical |